Molecule Information
General Information of the Molecule (ID: Mol01084)
| Name |
Solute carrier family 29 member 1 (SLC29A1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Equilibrative nitrobenzylmercaptopurine riboside-sensitive nucleoside transporter; Equilibrative NBMPR-sensitive nucleoside transporter; Nucleoside transporter; es-type; Solute carrier family 29 member 1; ENT1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
SLC29A1
|
||||
| Gene ID | |||||
| Location |
chr6:44219553-44234142[+]
|
||||
| Sequence |
MTTSHQPQDRYKAVWLIFFMLGLGTLLPWNFFMTATQYFTNRLDMSQNVSLVTAELSKDA
QASAAPAAPLPERNSLSAIFNNVMTLCAMLPLLLFTYLNSFLHQRIPQSVRILGSLVAIL LVFLITAILVKVQLDALPFFVITMIKIVLINSFGAILQGSLFGLAGLLPASYTAPIMSGQ GLAGFFASVAMICAIASGSELSESAFGYFITACAVIILTIICYLGLPRLEFYRYYQQLKL EGPGEQETKLDLISKGEEPRAGKEESGVSVSNSQPTNESHSIKAILKNISVLAFSVCFIF TITIGMFPAVTVEVKSSIAGSSTWERYFIPVSCFLTFNIFDWLGRSLTAVFMWPGKDSRW LPSLVLARLVFVPLLLLCNIKPRRYLTVVFEHDAWFIFFMAAFAFSNGYLASLCMCFGPK KVKPAEAETAGAIMAFFLCLGLALGAVFSFLFRAIV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Mediates both influx and efflux of nucleosides across the membrane (equilibrative transporter). It is sensitive (ES) to low concentrations of the inhibitor nitrobenzylmercaptopurine riboside (NBMPR) and is sodium-independent. It has a higher affinity for adenosine. Inhibited by dipyridamole and dilazep (anticancer chemotherapeutics drugs).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [1] | |||
| Resistant Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Resistant Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic ductal adenocarcinoma | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.44E-05 Fold-change: -6.19E-01 Z-score: -4.29E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Gemcitabine could be a substrate for several nucleoside transporters (NTs), but its major uptake occurs via the equilibrative and concentrative type NTs (ENTs and CNTs, respectively). ENT1, CNT1 and CNT3 have often been related to gemcitabine transport and resistance in humans. When ENT1 knockout conferred gemcitabine resistance, while its up regulation enhanced its cytotoxic activity. Similarly, retroviral expression of CNT1 renders ovarian cancer cells sensitive to gemcitabine in vitro. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [2] | |||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Resistant Drug | Cladribine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain | 1280 | ||
| Mechanism Description | Resistance of cytarabine resistant CEM cells to cladribine, among other purine and pyrimidine nucleoside drugs, was due to the absence of expression of the hENT1 gene leading to a loss ofnucleoside transport activity. Stable transfer of hCNT2 DNA into these resistant cells partially restored chemosensitivity for cladribine. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Leukemia [ICD-11: 2B33.6] | [3] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Resistant Drug | Cytarabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Cytarabine gains entry into cells primarily as a false substrate through specialized nucleoside transporter proteins of SLC family, the human equilibrative nucleoside transportershENT1 and hENT2 (encoded by the gene SLC29A1 and SCL29A2, respectively) and the human concentrative nucleoside transporters hCNT3 (encoded by the gene SLC28A3). The drug accumulation may be substantially reduced when the expression of hENT1 transporter is deficient, or the activity of ABC drug efflux transporter proteins is elevated. | |||
| Disease Class: Lymphoma [ICD-11: 2A90- 2A85] | [3] | |||
| Resistant Disease | Lymphoma [ICD-11: 2A90- 2A85] | |||
| Resistant Drug | Cytarabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Cytarabine gains entry into cells primarily as a false substrate through specialized nucleoside transporter proteins of SLC family, the human equilibrative nucleoside transportershENT1 and hENT2 (encoded by the gene SLC29A1 and SCL29A2, respectively) and the human concentrative nucleoside transporters hCNT3 (encoded by the gene SLC28A3). The drug accumulation may be substantially reduced when the expression of hENT1 transporter is deficient, or the activity of ABC drug efflux transporter proteins is elevated. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Tonsil tissue | |
| The Specified Disease | Lymphoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.60E-01; Fold-change: -7.50E-02; Z-score: -3.77E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
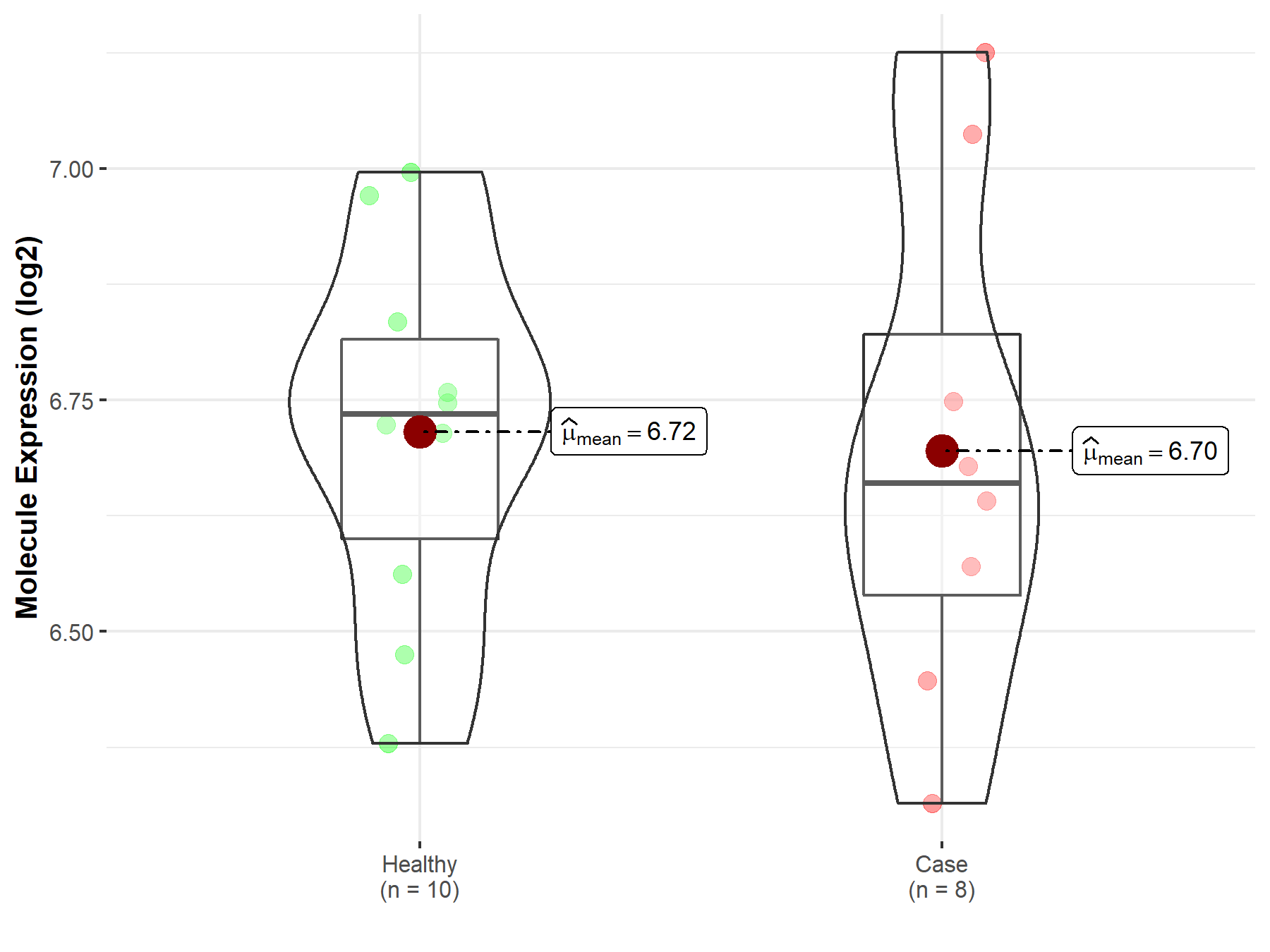
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.50E-01; Fold-change: 2.34E-01; Z-score: 5.51E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.83E-04; Fold-change: -5.18E-01; Z-score: -8.60E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
