Molecule Information
General Information of the Molecule (ID: Mol00655)
| Name |
Signal transducer activator transcription 3 (STAT3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Acute-phase response factor; APRF
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
STAT3
|
||||
| Gene ID | |||||
| Location |
chr17:42313324-42388540[-]
|
||||
| Sequence |
MAQWNQLQQLDTRYLEQLHQLYSDSFPMELRQFLAPWIESQDWAYAASKESHATLVFHNL
LGEIDQQYSRFLQESNVLYQHNLRRIKQFLQSRYLEKPMEIARIVARCLWEESRLLQTAA TAAQQGGQANHPTAAVVTEKQQMLEQHLQDVRKRVQDLEQKMKVVENLQDDFDFNYKTLK SQGDMQDLNGNNQSVTRQKMQQLEQMLTALDQMRRSIVSELAGLLSAMEYVQKTLTDEEL ADWKRRQQIACIGGPPNICLDRLENWITSLAESQLQTRQQIKKLEELQQKVSYKGDPIVQ HRPMLEERIVELFRNLMKSAFVVERQPCMPMHPDRPLVIKTGVQFTTKVRLLVKFPELNY QLKIKVCIDKDSGDVAALRGSRKFNILGTNTKVMNMEESNNGSLSAEFKHLTLREQRCGN GGRANCDASLIVTEELHLITFETEVYHQGLKIDLETHSLPVVVISNICQMPNAWASILWY NMLTNNPKNVNFFTKPPIGTWDQVAEVLSWQFSSTTKRGLSIEQLTTLAEKLLGPGVNYS GCQITWAKFCKENMAGKGFSFWVWLDNIIDLVKKYILALWNEGYIMGFISKERERAILST KPPGTFLLRFSESSKEGGVTFTWVEKDISGKTQIQSVEPYTKQQLNNMSFAEIIMGYKIM DATNILVSPLVYLYPDIPKEEAFGKYCRPESQEHPEADPGSAAPYLKTKFICVTPTTCSN TIDLPMSPRTLDSLMQFGNNGEGAEPSAGGQFESLTFDMELTSECATSPM Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Signal transducer and transcription activator that mediates cellular responses to interleukins, KITLG/SCF, LEP and other growth factors. Once activated, recruits coactivators, such as NCOA1 or MED1, to the promoter region of the target gene. May mediate cellular responses to activated FGFR1, FGFR2, FGFR3 and FGFR4. Upon activation of IL6ST/gp130 signaling by interleukin-6 (IL6), binds to the IL6-responsive elements identified in the promoters of various acute-phase protein genes. Activated by IL31 through IL31RA. Acts as a regulator of inflammatory response by regulating differentiation of naive CD4(+) T-cells into T-helper Th17 or regulatory T-cells (Treg): deacetylation and oxidation of lysine residues by LOXL3, leads to disrupt STAT3 dimerization and inhibit its transcription activity. Involved in cell cycle regulation by inducing the expression of key genes for the progression from G1 to S phase, such as CCND1. Mediates the effects of LEP on melanocortin production, body energy homeostasis and lactation. May play an apoptotic role by transctivating BIRC5 expression under LEP activation. Cytoplasmic STAT3 represses macroautophagy by inhibiting EIF2AK2/PKR activity. Plays a crucial role in basal beta cell functions, such as regulation of insulin secretion.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
14 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [1] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Taxane | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.76E-01 Fold-change: -2.32E-02 Z-score: -8.88E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H1155 cells | Lung | Homo sapiens (Human) | CVCL_1456 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Over-expression of miR-337-3p nor the specific knockdown of STAT3 and RAP1A significantly decrease cell viability or induce G2/M arrest alone, but rather enhance G2/M arrest and cell death only under conditions of paclitaxel treatment. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [2] | |||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung adenocarcinoma | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.05E-02 Fold-change: 6.61E-02 Z-score: 2.60E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell invasion | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via upregulating Stat3 by sequestering miR-454-3p. | |||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [4], [12] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell cycle | Activation | hsa04110 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3k/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR34a. | |||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [13] | |||
| Resistant Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Siha cells | Cervix uteri | Homo sapiens (Human) | CVCL_0032 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Colony formation assay | |||
| Mechanism Description | Down-regulation of LncRNA GAS5 strengthen cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [4] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Hepatocellular carcinoma | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.12E-04 Fold-change: -1.39E-01 Z-score: -3.93E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3/ABCB1 signaling pathway | Inhibition | hsa05200 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3k/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR34a. | |||
| Disease Class: Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | [14] | |||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| STAT3 signaling pathway | Inhibition | hsa04550 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 | |
| EC9706 cells | Esophagus | Homo sapiens (Human) | CVCL_E307 | |
| EC1 cells | Esophagus | Homo sapiens (Human) | CVCL_DC74 | |
| KYSE70 cells | Esophagus | Homo sapiens (Human) | CVCL_1356 | |
| kYSE450 cells | Esophagus | Homo sapiens (Human) | CVCL_1353 | |
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Would healing assay; Invasion assay | |||
| Mechanism Description | miR 125a 5p and cisplatin markedly inactivated the STAT3 signaling pathway. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [3] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.76E-01 Fold-change: -2.32E-02 Z-score: -8.88E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase Assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-FITC Apoptosis assay | |||
| Mechanism Description | miR124 decreased SNAI2 and STAT3 expression by directly targeting their 3'UTRs, miR124 contributes to gefitinib and EMT by directly targeting SNAI2 and STAT3. Over-expression of miR124 re-sensitized gefitinib-resistant cell lines to gefitinib. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [20] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| STAT3 signaling pathway | Inhibition | hsa04550 | ||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | PPI might down-regulate MALAT1 expression and inactivate STAT3 signaling pathway and could serve a promising therapeutic agent for gefitinib-resistant NSCLC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal cell carcinoma [ICD-11: 2C90.0] | [6] | |||
| Resistant Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Resistant Drug | Sorafenib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cancer | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.66E-01 Fold-change: 1.15E-02 Z-score: 4.44E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Sorafenib tolerance | Activation | hsa00983 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| A498 cells | Kidney | Homo sapiens (Human) | CVCL_1056 | |
| Caki-2 cells | Kidney | Homo sapiens (Human) | CVCL_0235 | |
| OSRC-2 cells | Kidney | Homo sapiens (Human) | CVCL_1626 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Long noncoding RNA-SRLR elicits intrinsic sorafenib resistance via evoking IL-6/STAT3 axis in renal cell carcinoma. LncRNA-SRLR directly binds to NF-kB and promotes IL-6 transcription, leading to the activation of STAT3 and the development of sorafenib tolerance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [25] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Sorafenib | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| STAT3 signaling pathway | Inhibition | hsa04550 | ||
| In Vitro Model | HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| In Vivo Model | NOD-SCID mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Interference lncARSR suppressed liver CSCs expansion and the phosphorylation of the STAT3 molecule was evidently inactivated in both the SMMC7721 si-lncARSR and HCCLM3 si-lncARSR cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [1] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.92E-04 Fold-change: -1.90E-02 Z-score: -3.57E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H1155 cells | Lung | Homo sapiens (Human) | CVCL_1456 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Over-expression of miR-337-3p nor the specific knockdown of STAT3 and RAP1A significantly decrease cell viability or induce G2/M arrest alone, but rather enhance G2/M arrest and cell death only under conditions of paclitaxel treatment. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [24] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3/p53 pathway | Inhibition | hsa05212 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
TUNEL assaay; Sulphorhodamine B assay | |||
| Mechanism Description | miR17-5p promoted apoptosis by increasing p53 expression, which was inhibited by STAT3, miR17-5p inhibits STAT3 and increases p53 expression to promote apoptosis in breast cancer cells. miR17-5p directly targets STAT3 and induces apoptosis in breast cancer cells by inhibiting the STAT3/p53 pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [11] | |||
| Resistant Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Phosphorylation | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SKOV-3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| OVCAR5 cells | Ovary | Homo sapiens (Human) | CVCL_1628 | |
| OVCAR8 cells | Ovary | Homo sapiens (Human) | CVCL_1629 | |
| Mechanism Description | We show that the addition of AAFs to the culture media of EOC cell lines has the potential to induce resistance to standard-of-care drugs (SCDs). We also show that AAFs induce time- and concentration-dependent activation of downstream signalling to signal transducer and activator of transcription 3 (STAT3), and concomitantly altered phosphorylation of mitogen-activated protein kinase kinase (MEK), phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT) and nuclear factor NF-kappa-B (NFkappaB). Antibodies targeting the interleukin-6 receptor (IL6R) effectively blocked phosphorylation of STAT3 and STAT1. | |||
|
|
||||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [23] | |||
| Resistant Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| JAKT/STAT signaling pathway | Regulation | N.A. | ||
| In Vitro Model | NP69 cells | Nasopharynx | Homo sapiens (Human) | CVCL_F755 |
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-29a down-regulation is correlated with drug resistance of nasopharyngeal carcinoma cell line CNE-1 and miR-29a up-regulation decreases Taxol resistance of nasopharyngeal carcinoma CNE-1 cells possibly via inhibiting STAT3 and Bcl-2 expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [7] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Cetuximab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.50E-03 Fold-change: -4.36E-02 Z-score: -2.83E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SNU449 cells | Liver | Homo sapiens (Human) | CVCL_0454 | |
| SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Let-7a enhances the sensitivity of hepatocellular carcinoma cells to cetuximab by negatively regulating STAT3 expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Corona Virus Disease 2019 [ICD-11: 1D92.0] | [10] | |||
| Sensitive Disease | Corona Virus Disease 2019 [ICD-11: 1D92.0] | |||
| Sensitive Drug | 6-N-ethyl-netilmicin | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Immune cells | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Flow cytometry | |||
| Mechanism Description | Baricitinib is a selective Janus kinase (JAK)1/JAK2 inhibitor with a known anti-inflammatory profile in patients with autoimmune diseases.Baricitinib was also shown to reduce multiple cytokines and biomarkers implicated in COVID-19 pathophysiology. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [11] | |||
| Resistant Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Resistant Drug | Carboplatin | |||
| Molecule Alteration | Phosphorylation | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SKOV-3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| OVCAR5 cells | Ovary | Homo sapiens (Human) | CVCL_1628 | |
| OVCAR8 cells | Ovary | Homo sapiens (Human) | CVCL_1629 | |
| Mechanism Description | We show that the addition of AAFs to the culture media of EOC cell lines has the potential to induce resistance to standard-of-care drugs (SCDs). We also show that AAFs induce time- and concentration-dependent activation of downstream signalling to signal transducer and activator of transcription 3 (STAT3), and concomitantly altered phosphorylation of mitogen-activated protein kinase kinase (MEK), phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT) and nuclear factor NF-kappa-B (NFkappaB). Antibodies targeting the interleukin-6 receptor (IL6R) effectively blocked phosphorylation of STAT3 and STAT1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [15] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Dacomitinib | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | EGFR signaling pathway | Inhibition | hsa01521 | |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
LDH assay; Flow cytometry assay | |||
| Mechanism Description | Our study aimed to analyze the cellular mechanism of dacomitinib, a pan-epidermal growth factor receptor (EGFR) inhibitor, which resensitized paclitaxel and induced cell cytotoxicity in paclitaxel-resistant ovarian cancer SKOV3-TR cells. We investigated the significant reduction in cell viability cotreated with dacomitinib and paclitaxel by WST-1 assay and flow cytometry analysis. Dacomitinib inhibited EGFR family proteins, including EGFR and HER2, as well as its downstream signaling proteins, including AKT, STAT3, ERK, and p38. In addition, dacomitinib inhibited the phosphorylation of Bad, and combination treatment with paclitaxel effectively suppressed the expression of Mcl-1. A 2'-7'-dichlorodihydrofluorescein diacetate (DCFH-DA) assay revealed a substantial elevation in cellular reactive oxygen species (ROS) levels in SKOV3-TR cells cotreated with dacomitinib and paclitaxel, which subsequently mediated cell cytotoxicity. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer bone metastasis [ICD-11: 2E03.1] | [16] | |||
| Resistant Disease | Breast cancer bone metastasis [ICD-11: 2E03.1] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometric | |||
| Mechanism Description | Interleukin-6 (IL-6), a pro-inflammatory cytokine produced in the tumor microenvironment by stromal cells, fibroblasts, and cancer cells. Binding of IL-6 to its receptor IL-6R on the cell membrane activates Janus Kinase 2 (JAK2) kinases. Activated JAK2 mediates phosphorylation, dimerization, and nuclear translocation of Signal Transducer and Activator of Transcription 3 (STAT3). STAT3 signaling mediates the expression of various genes, including p53, Bcl-2, MRP1, and ABCG2. Bcl-2 and p53 are associated with regulation of apoptosis while overexpression of drug transporters MRP1, ABCG2 has been shown to mediate efflux of drugs from cancer cells, thus decreasing intracellular drug concentration leading to drug-resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3/HIF-1 signaling pathway | Inhibition | hsa04066 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 sensitizes DOX-resistant BCSCs to DOX by modulating the STAT3 and HIF-1 signaling pathways. | |||
| Disease Class: Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | [18] | |||
| Sensitive Disease | Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| STAT3/INO80 signaling pathway | Inhibition | hsa04066 | ||
| In Vitro Model | FTC-133 cells | Thyroid | Homo sapiens (Human) | CVCL_1219 |
| 8505C cells | Thyroid | Homo sapiens (Human) | CVCL_1054 | |
| FTC 238 cells | Thyroid | Homo sapiens (Human) | CVCL_2447 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | LncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [19] | |||
| Resistant Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Resistant Drug | Everolimus | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Activation | hsa04218 | |
| In Vitro Model | QGP-1R cells | carcinoid | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Cell viability assay; Colony formation assay | |||
| Mechanism Description | The activation of the STAT3 pathway induced by TNF is mediated by NF-kB p65. NF-kB p65 and STAT3 inhibitors decrease QGP-1 viability, spheroids growth, and Pa-NETs cell proliferation. These effects are maintained in everolimus-resistant QGP-1R cells. | |||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [19] | |||
| Resistant Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Resistant Drug | Everolimus | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Activation | hsa04218 | |
| In Vitro Model | QGP-1R cells | carcinoid | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Cell viability assay; Colony formation assay | |||
| Mechanism Description | The activation of the STAT3 pathway induced by TNF is mediated by NF-kB p65. NF-kB p65 and STAT3 inhibitors decrease QGP-1 viability, spheroids growth, and Pa-NETs cell proliferation. These effects are maintained in everolimus-resistant QGP-1R cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Extensive-disease small-cell lung cancer [ICD-11: 2C25.Z] | [21] | |||
| Sensitive Disease | Extensive-disease small-cell lung cancer [ICD-11: 2C25.Z] | |||
| Sensitive Drug | Ipilimumab | |||
| Molecule Alteration | Function | Inhibition |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Drug Resistance |
FoundationOne CDx assay | |||
| Mechanism Description | Overcoming acquired resistance to chemotherapy treatments through suppression of STAT3, Selective chemotherapy treatments and diagnostic methods related thereto, DR4 modulation and its implications in EGFR-target cancer therapy. Maintenance therapy with nivolumab plus ipilimumab did not prolong OS for patients with ED-SCLC who did not progress on first-line chemotherapy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [22] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Ivermectin | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | EGFR/STAT3/ERK signalling pathway | Regulation | N.A. | |
| In Vitro Model | K562/FLM cells | Blood | Homo sapiens (Human) | CVCL_E7CM |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | It was found that ivermectin effectively suppressed the expression of autophagy and transport proteins in K562/FLM cells, reduced the activity of the aforementioned phosphoproteins, and promoted apoptotic cell death. The significant effects of ivermectin might offer a novel therapeutic strategy to overcome flumatinib resistance and optimize the treatment outcomes of CML. | |||
Clinical Trial Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [5] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cucurbitacin B | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.27E-01 Fold-change: 9.64E-02 Z-score: 2.52E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | GACAT3 alleviates the anticancer drug cucurbitacin B-induced apoptosis of gastric cancer cells via increasing STAT3 expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [19] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | AZD1480 | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Activation | hsa04218 | |
| In Vitro Model | QGP-1R cells | carcinoid | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Cell viability assay; Colony formation assay | |||
| Mechanism Description | The activation of the STAT3 pathway induced by TNF is mediated by NF-kB p65. NF-kB p65 and STAT3 inhibitors decrease QGP-1 viability, spheroids growth, and Pa-NETs cell proliferation. These effects are maintained in everolimus-resistant QGP-1R cells. | |||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [19] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | AZD1480 | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Activation | hsa04218 | |
| In Vitro Model | QGP-1R cells | carcinoid | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Cell viability assay; Colony formation assay | |||
| Mechanism Description | The activation of the STAT3 pathway induced by TNF is mediated by NF-kB p65. NF-kB p65 and STAT3 inhibitors decrease QGP-1 viability, spheroids growth, and Pa-NETs cell proliferation. These effects are maintained in everolimus-resistant QGP-1R cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [22] | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Resistant Drug | Flumatinib | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | EGFR/STAT3/ERK signalling pathway | Regulation | N.A. | |
| In Vitro Model | K562/FLM cells | Blood | Homo sapiens (Human) | CVCL_E7CM |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Through cellular experimentation, we explored the resistance mechanisms, which indicated that K562/FLM cells evade flumatinib cytotoxicity by enhancing autophagy, increasing the expression of membrane transport proteins, particularly P-glycoprotein, ABCC1 and ABCC4, as well as enhancing phosphorylation of p-EGFR, p-ERK and p-STAT3 proteins. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Esophagus | |
| The Specified Disease | Esophageal cancer | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.40E-03; Fold-change: -8.87E-01; Z-score: -2.52E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.27E-01; Fold-change: 6.53E-01; Z-score: 1.61E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.16E-10; Fold-change: 6.83E-01; Z-score: 3.20E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.50E-03; Fold-change: -1.16E-01; Z-score: -2.11E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.09E-01; Fold-change: 1.87E-01; Z-score: 3.01E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 8.64E-02; Fold-change: -5.60E-01; Z-score: -1.07E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
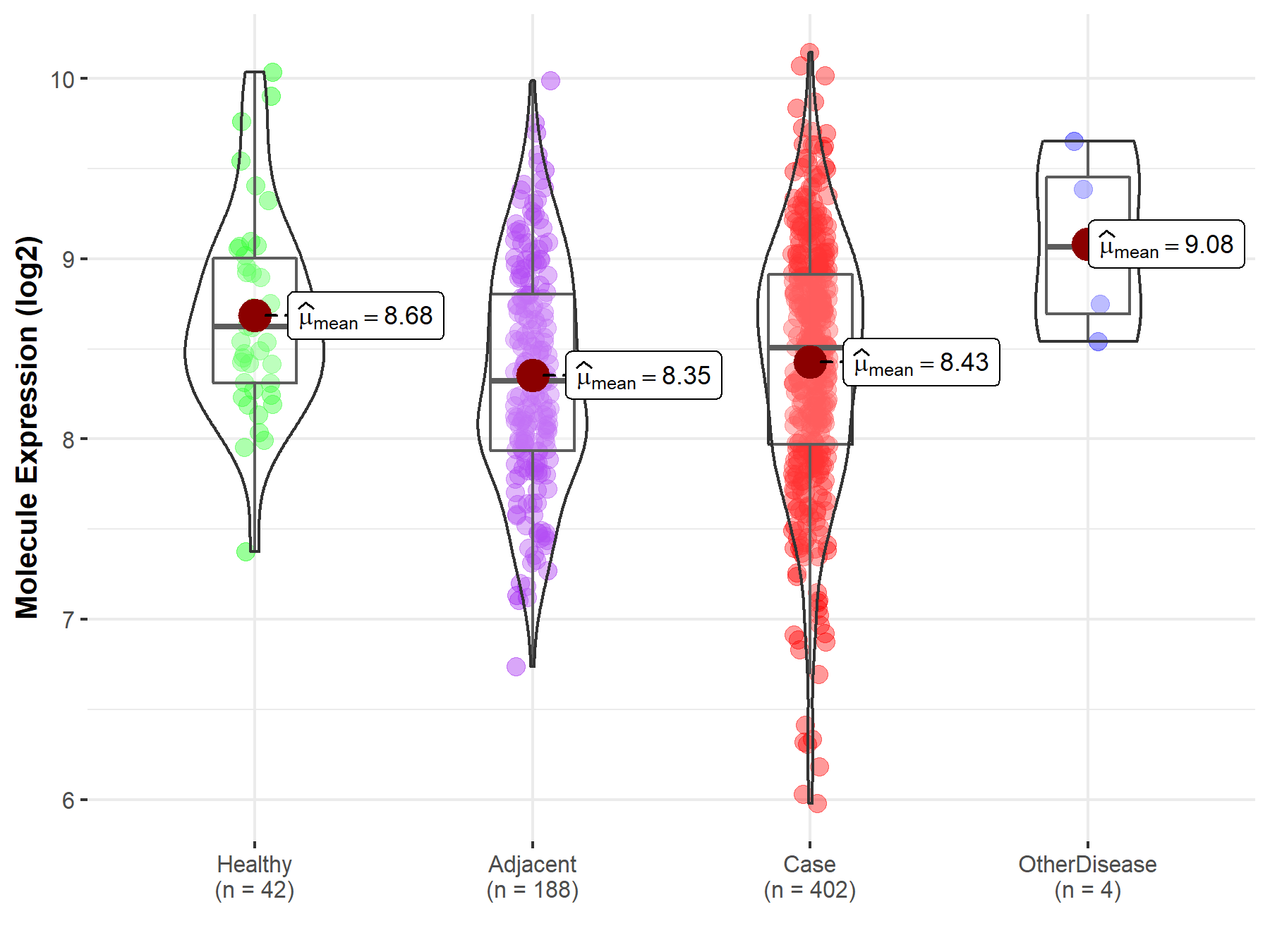
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.92E-04; Fold-change: -5.85E-02; Z-score: -1.58E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.99E-04; Fold-change: -2.42E-01; Z-score: -4.08E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
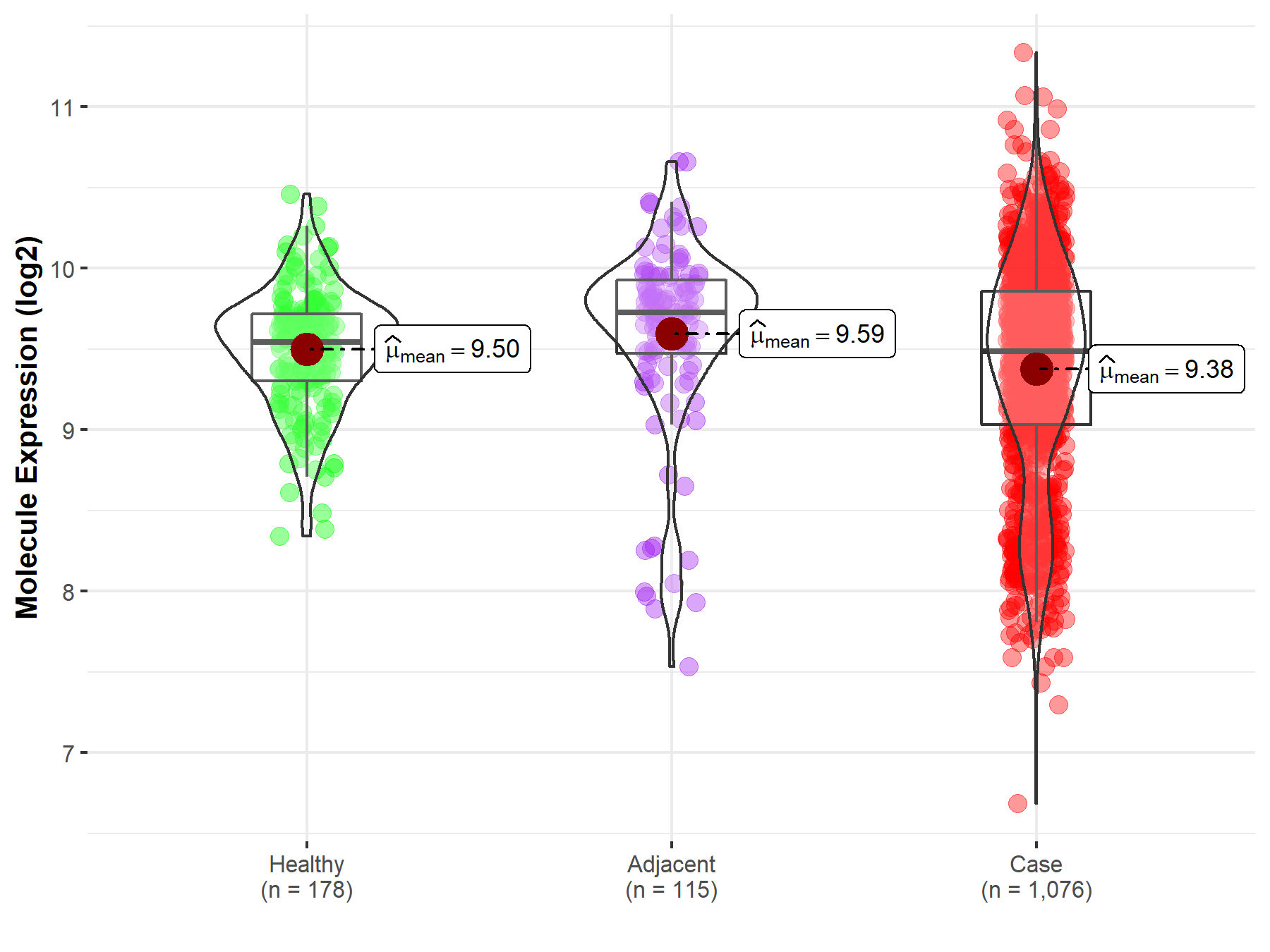
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.18E-01; Fold-change: 1.31E-01; Z-score: 1.69E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.31E-02; Fold-change: 6.75E-02; Z-score: 1.04E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.71E-03; Fold-change: 1.73E-01; Z-score: 5.67E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
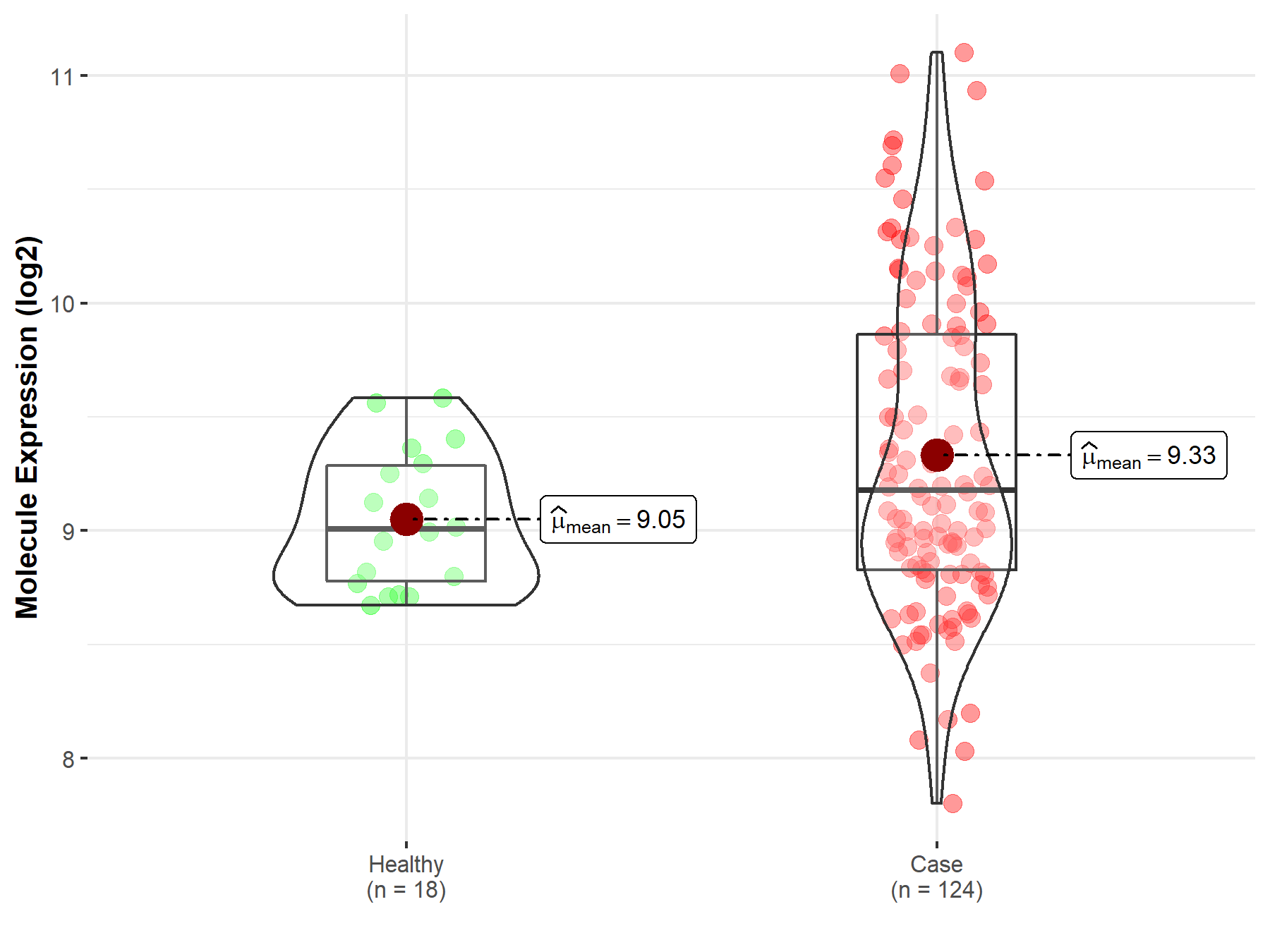
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Kidney | |
| The Specified Disease | Kidney cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.66E-01; Fold-change: 1.26E-01; Z-score: 2.43E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.07E-07; Fold-change: -1.18E-01; Z-score: -3.62E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
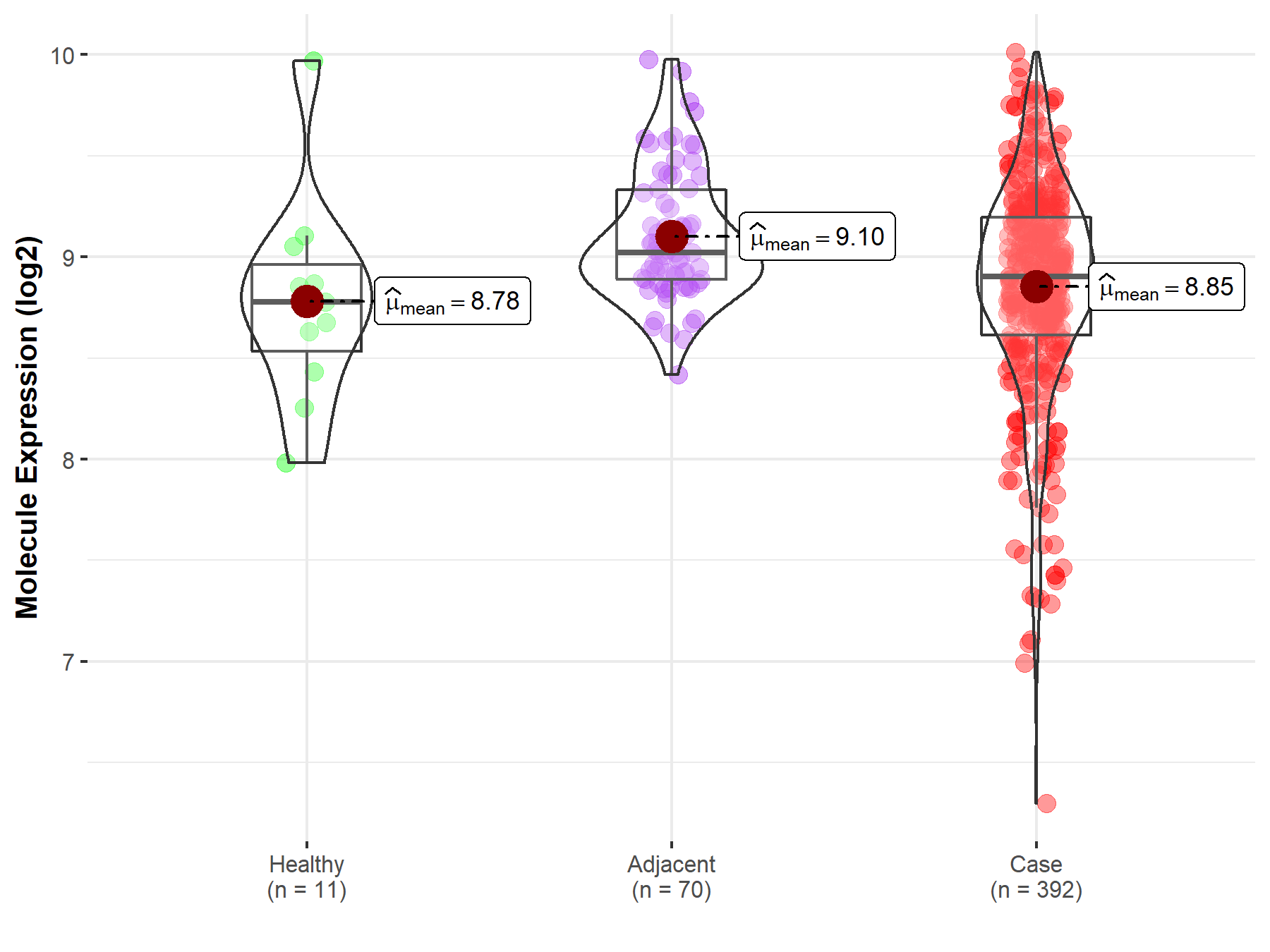
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.80E-03; Fold-change: 2.44E-01; Z-score: 6.98E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.03E-08; Fold-change: 6.57E-01; Z-score: 1.24E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
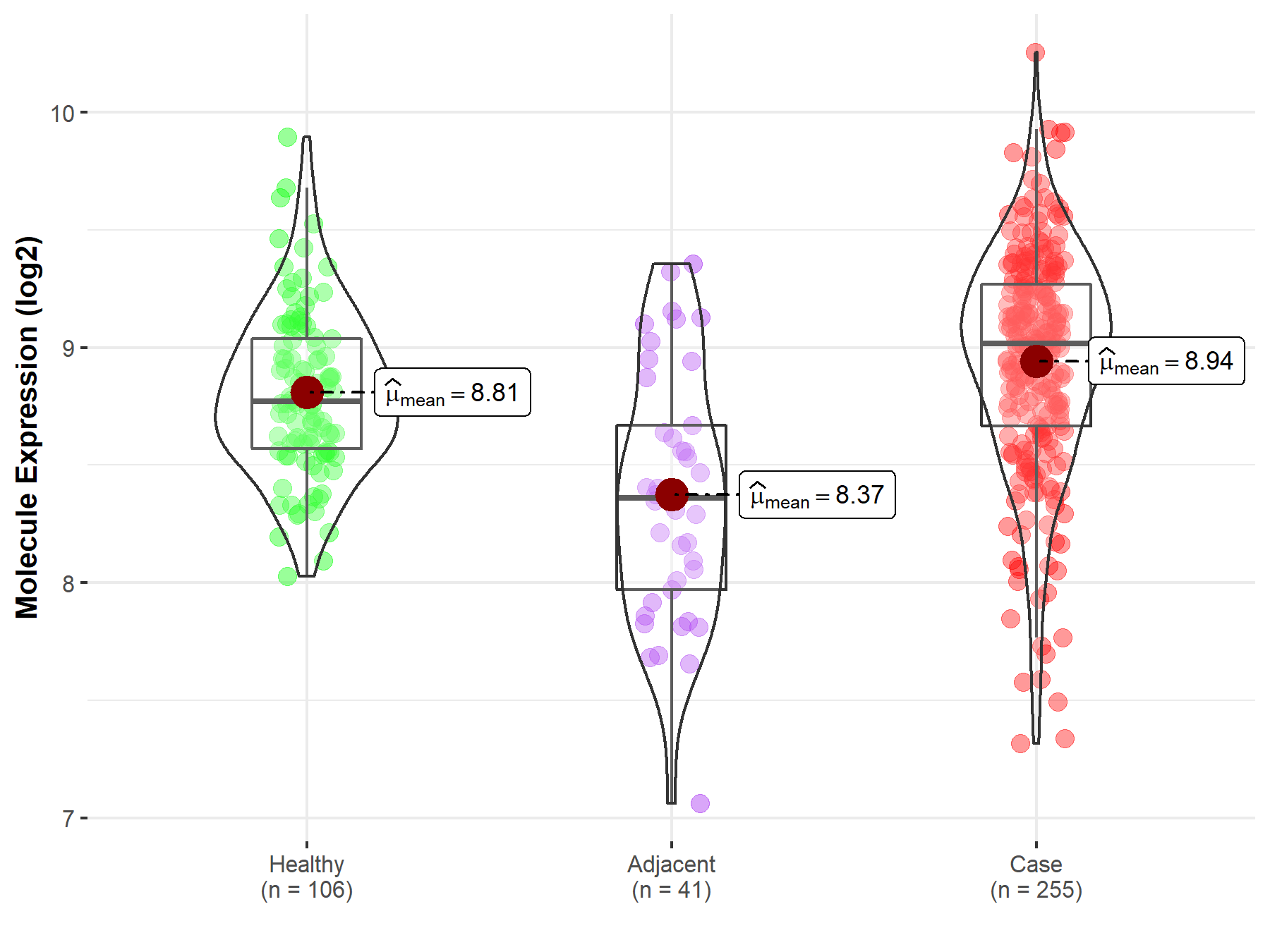
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

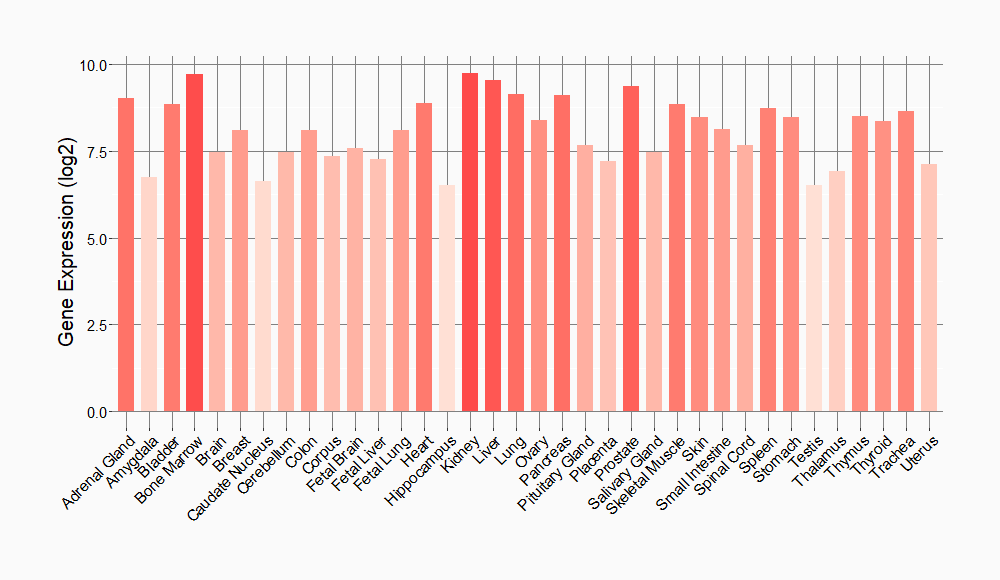
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
