Molecule Information
General Information of the Molecule (ID: Mol00331)
| Name |
Dihydropyrimidine dehydrogenase [NADP(+)]
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
DHPDHase; DPD; Dihydrothymine dehydrogenase; Dihydrouracil dehydrogenase
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
DPYD
|
||||
| Gene ID | |||||
| Location |
chr1:97077743-97995000[-]
|
||||
| Sequence |
MAPVLSKDSADIESILALNPRTQTHATLCSTSAKKLDKKHWKRNPDKNCFNCEKLENNFD
DIKHTTLGERGALREAMRCLKCADAPCQKSCPTNLDIKSFITSIANKNYYGAAKMIFSDN PLGLTCGMVCPTSDLCVGGCNLYATEEGPINIGGLQQFATEVFKAMSIPQIRNPSLPPPE KMSEAYSAKIALFGAGPASISCASFLARLGYSDITIFEKQEYVGGLSTSEIPQFRLPYDV VNFEIELMKDLGVKIICGKSLSVNEMTLSTLKEKGYKAAFIGIGLPEPNKDAIFQGLTQD QGFYTSKDFLPLVAKGSKAGMCACHSPLPSIRGVVIVLGAGDTAFDCATSALRCGARRVF IVFRKGFVNIRAVPEEMELAKEEKCEFLPFLSPRKVIVKGGRIVAMQFVRTEQDETGKWN EDEDQMVHLKADVVISAFGSVLSDPKVKEALSPIKFNRWGLPEVDPETMQTSEAWVFAGG DVVGLANTTVESVNDGKQASWYIHKYVQSQYGASVSAKPELPLFYTPIDLVDISVEMAGL KFINPFGLASATPATSTSMIRRAFEAGWGFALTKTFSLDKDIVTNVSPRIIRGTTSGPMY GPGQSSFLNIELISEKTAAYWCQSVTELKADFPDNIVIASIMCSYNKNDWTELAKKSEDS GADALELNLSCPHGMGERGMGLACGQDPELVRNICRWVRQAVQIPFFAKLTPNVTDIVSI ARAAKEGGANGVTATNTVSGLMGLKSDGTPWPAVGIAKRTTYGGVSGTAIRPIALRAVTS IARALPGFPILATGGIDSAESGLQFLHSGASVLQVCSAIQNQDFTVIEDYCTGLKALLYL KSIEELQDWDGQSPATVSHQKGKPVPRIAELMDKKLPSFGPYLEQRKKIIAENKIRLKEQ NVAFSPLKRNCFIPKRPIPTIKDVIGKALQYLGTFGELSNVEQVVAMIDEEMCINCGKCY MTCNDSGYQAIQFDPETHLPTITDTCTGCTLCLSVCPIVDCIKMVSRTTPYEPKRGVPLS VNPVC Click to Show/Hide
|
||||
| Function |
Involved in pyrimidine base degradation. Catalyzes the reduction of uracil and thymine. Also involved the degradation of the chemotherapeutic drug 5-fluorouracil.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.42E-01 Fold-change: 5.46E-03 Z-score: 6.11E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | |
| NCI-H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| PC-10 cells | Lung | Homo sapiens (Human) | CVCL_7088 | |
| QG56 cells | Lung | Homo sapiens (Human) | CVCL_6943 | |
| RERF-LCMS cells | Lung | Homo sapiens (Human) | CVCL_1655 | |
| ACC-LC-176 cells | Lung | Homo sapiens (Human) | CVCL_7008 | |
| RERF-LC-MT cells | Lung | Homo sapiens (Human) | CVCL_A473 | |
| RERF-LC-Ok cell | Lung | Homo sapiens (Human) | CVCL_3154 | |
| Sk-LC-10 cells | Lung | Homo sapiens (Human) | CVCL_5459 | |
| Sk-LC-6 cells | Lung | Homo sapiens (Human) | CVCL_5474 | |
| VMRC-LCD cells | Lung | Homo sapiens (Human) | CVCL_1787 | |
| VMRC-LCF cells | Lung | Homo sapiens (Human) | CVCL_S848 | |
| Experiment for Molecule Alteration |
PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Degradation of 5-FU due to DPD is an important determinant in 5-FU sensitivity, while induction of TS contributes to acquired resistance against 5-FU in lung cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [2] | |||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-04 Fold-change: -2.76E-02 Z-score: -3.90E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | 5-Fu catabolic signaling pathway | Regulation | N.A. | |
| Cell apoptosis | Activation | hsa04210 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| HCT15 cells | Colon | Homo sapiens (Human) | CVCL_0292 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-494 also negatively regulated endogenous DPYD expression in SW480 cells. Overexpression or knockdown of DPYD could attenuate miR-494 mediated 5-Fu sensitivity regulation, suggesting the dependence of DPYD regulation in miR-494 activity. miR-494 inhibited SW480/5-Fu derived xenograft tumors growth in vivo at present of 5-Fu. | |||
| Disease Class: Esophageal cancer [ICD-11: 2B70.1] | [3] | |||
| Sensitive Disease | Esophageal cancer [ICD-11: 2B70.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Methylation | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 | |
| KYSE150 cells | Esophagus | Homo sapiens (Human) | CVCL_1348 | |
| TE-5 cells | Esophageal | Homo sapiens (Human) | CVCL_1764 | |
| In Vivo Model | BALB/c nude mouse xenograft mode | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 assay; Flow cytometry assay | |||
| Mechanism Description | Long noncoding RNA LINC00261 induces chemosensitization to 5-fluorouracil by mediating methylation-dependent repression of DPYD in human esophageal cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Tegafur | |||
| Molecule Alteration | Missense mutation | p.I560S (c.1679T>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Tegafur | |||
| Molecule Alteration | Missense mutation | p.D949V (c.2846A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Esophagus | |
| The Specified Disease | Esophageal cancer | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.18E-02; Fold-change: -3.45E-01; Z-score: -1.12E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-04; Fold-change: -8.60E-02; Z-score: -3.50E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.01E-01; Fold-change: -3.00E-02; Z-score: -1.30E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
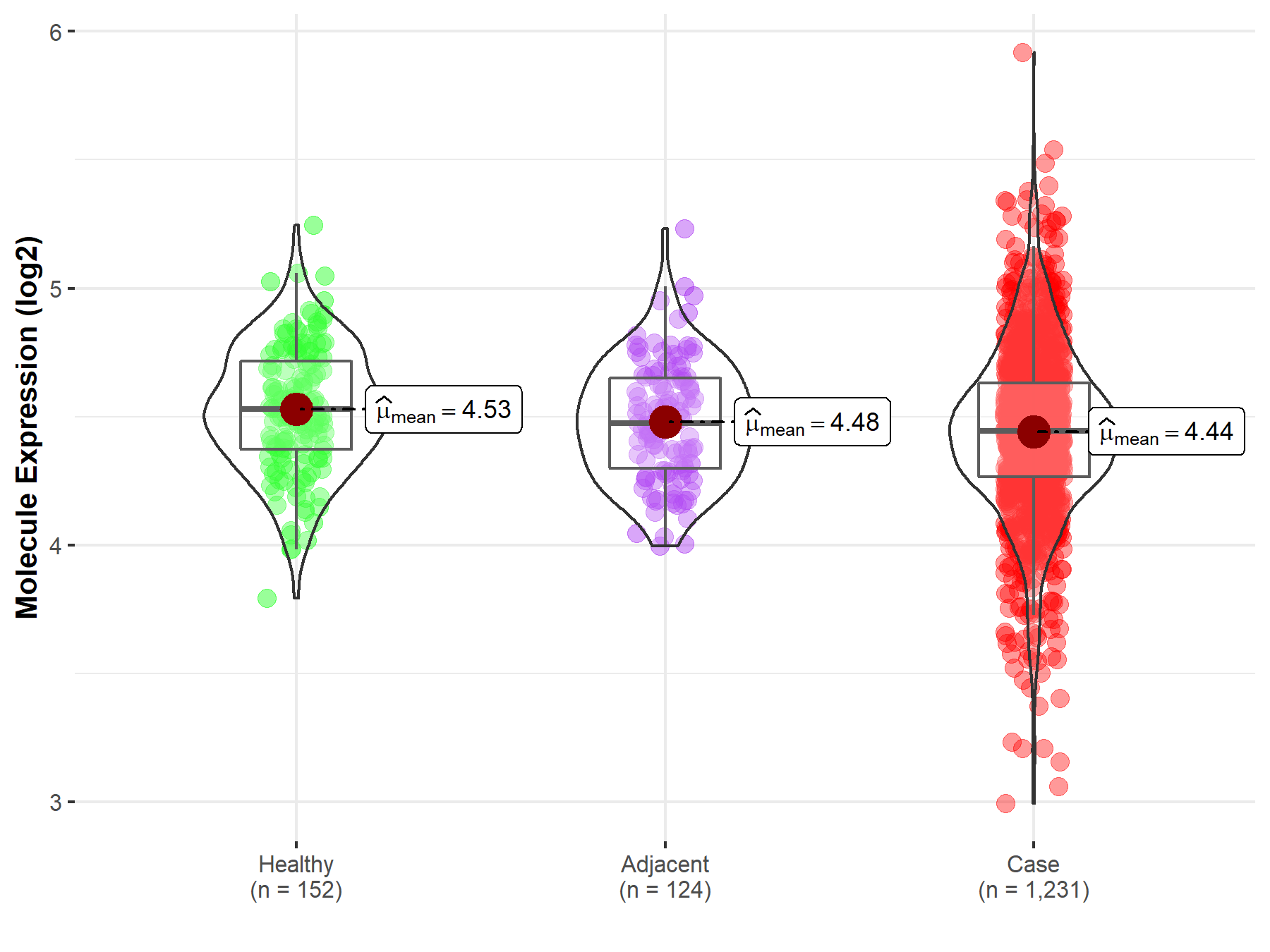
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.42E-01; Fold-change: -7.54E-02; Z-score: -2.16E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.62E-01; Fold-change: -8.61E-02; Z-score: -2.54E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
