Molecule Information
General Information of the Molecule (ID: Mol00196)
| Name |
Zinc finger E-box-binding homeobox 2 (ZEB2)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Smad-interacting protein 1; SMADIP1; Zinc finger homeobox protein 1b; KIAA0569; SIP1; ZFHX1B; ZFX1B; HRIHFB2411
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ZEB2
|
||||
| Gene ID | |||||
| Location |
chr2:144364364-144521057[-]
|
||||
| Sequence |
MKQPIMADGPRCKRRKQANPRRKNVVNYDNVVDTGSETDEEDKLHIAEDDGIANPLDQET
SPASVPNHESSPHVSQALLPREEEEDEIREGGVEHPWHNNEILQASVDGPEEMKEDYDTM GPEATIQTAINNGTVKNANCTSDFEEYFAKRKLEERDGHAVSIEEYLQRSDTAIIYPEAP EELSRLGTPEANGQEENDLPPGTPDAFAQLLTCPYCDRGYKRLTSLKEHIKYRHEKNEEN FSCPLCSYTFAYRTQLERHMVTHKPGTDQHQMLTQGAGNRKFKCTECGKAFKYKHHLKEH LRIHSGEKPYECPNCKKRFSHSGSYSSHISSKKCIGLISVNGRMRNNIKTGSSPNSVSSS PTNSAITQLRNKLENGKPLSMSEQTGLLKIKTEPLDFNDYKVLMATHGFSGTSPFMNGGL GATSPLGVHPSAQSPMQHLGVGMEAPLLGFPTMNSNLSEVQKVLQIVDNTVSRQKMDCKA EEISKLKGYHMKDPCSQPEEQGVTSPNIPPVGLPVVSHNGATKSIIDYTLEKVNEAKACL QSLTTDSRRQISNIKKEKLRTLIDLVTDDKMIENHNISTPFSCQFCKESFPGPIPLHQHE RYLCKMNEEIKAVLQPHENIVPNKAGVFVDNKALLLSSVLSEKGMTSPINPYKDHMSVLK AYYAMNMEPNSDELLKISIAVGLPQEFVKEWFEQRKVYQYSNSRSPSLERSSKPLAPNSN PPTKDSLLPRSPVKPMDSITSPSIAELHNSVTNCDPPLRLTKPSHFTNIKPVEKLDHSRS NTPSPLNLSSTSSKNSHSSSYTPNSFSSEELQAEPLDLSLPKQMKEPKSIIATKNKTKAS SISLDHNSVSSSSENSDEPLNLTFIKKEFSNSNNLDNKSTNPVFSMNPFSAKPLYTALPP QSAFPPATFMPPVQTSIPGLRPYPGLDQMSFLPHMAYTYPTGAATFADMQQRRKYQRKQG FQGELLDGAQDYMSGLDDMTDSDSCLSRKKIKKTESGMYACDLCDKTFQKSSSLLRHKYE HTGKRPHQCQICKKAFKHKHHLIEHSRLHSGEKPYQCDKCGKRFSHSGSYSQHMNHRYSY CKREAEEREAAEREAREKGHLEPTELLMNRAYLQSITPQGYSDSEERESMPRDGESEKEH EKEGEDGYGKLGRQDGDEEFEEEEEESENKSMDTDPETIRDEEETGDHSMDDSSEDGKME TKSDHEEDNMEDGM Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Transcriptional inhibitor that binds to DNA sequence 5'-CACCT-3' in different promoters. Represses transcription of E-cadherin. Represses expression of MEOX2.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Epothilone B | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-35 Fold-change: -9.00E-01 Z-score: -1.66E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-35 Fold-change: -9.00E-01 Z-score: -1.66E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-35 Fold-change: -9.00E-01 Z-score: -1.66E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.32E-22 Fold-change: -4.94E-01 Z-score: -1.11E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| ERK signaling pathway | Regulation | N.A. | ||
| RhoC/FAKT/Src/ROCK2 signaling pathways | Regulation | N.A. | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| NCI-H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-138 sensitizes NSCLC cells to ADM through regulation of EMT regulator ZEB2. Ectopic expression of miR-138 decreased ZEB2 expression and inhibited the luciferase activity in chemoresistant tumor cells, suggesting that miR-138 could regulate EMT, at least partly, through targeting ZEB2 in NSCLC cells. | |||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.51E-22 Fold-change: -1.97E-01 Z-score: -1.05E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [3] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung adenocarcinoma | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.32E-22 Fold-change: -4.94E-01 Z-score: -1.11E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Inhibition | hsa01521 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Direct interaction between miR203 and ZEB2 suppresses epithelial-mesenchymal transition signaling and reduces lung adenocarcinoma chemoresistance. ZEB2 was found to be a direct target of miR203, which induces epithelial-mesenchymal transition (EMT) signal. | |||
| Disease Class: Lung small cell carcinoma [ICD-11: 2C25.2] | [5] | |||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.68E-67 Fold-change: -2.33E-01 Z-score: -2.10E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-200b was down-regulated in the resistant cells and enforced expression of miR-200b by miRNA mimics increased cell sensitivity. Overexpression of miR-200b led to the downregulation of ZEB2 at protein level. Luciferase reporter gene assay showed that 3'UTR ZEB2 activity was regulated by miR-200b. ZEB2 modulates drug resistance and is regulated by miR-200b. knockdown of ZEB2 increased cell sensitivity through increasing drug-induced cell apoptosis accompanied with S phase arrest. ZEB2 was regulated by miR-200b at protein level. | |||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [7] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ZEB2 signaling pathway | Inhibition | hsa05202 | |
| In Vitro Model | SGC7901/DDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Dual luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. | |||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [8] | |||
| Sensitive Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | 5-8F cells | Nasopharynx | Homo sapiens (Human) | CVCL_C528 |
| 6-10B cells | Nasopharynx | Homo sapiens (Human) | CVCL_C529 | |
| SUNE-1 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6946 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | There is a directly negative feedback loop between miR203 and ZEB2 participating in tumor stemness and chemotherapy resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [6] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Missense mutation | p.Y663C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The miR205-5p and miR200 families can silence ZEB1 and ZEB2 expression. LncRNA-ROR functions as a molecular sponge for miR205-5p and affects the target genes ZEB1 and ZEB2, which in turn influences the EMT process in breast cancer cells. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.37E-01; Fold-change: -6.12E-02; Z-score: -1.88E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.81E-01; Fold-change: -2.39E-01; Z-score: -5.08E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.68E-67; Fold-change: -9.50E-01; Z-score: -1.93E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.78E-39; Fold-change: -1.01E+00; Z-score: -1.69E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.51E-22; Fold-change: -9.55E-01; Z-score: -8.00E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.54E-04; Fold-change: -5.20E-01; Z-score: -4.41E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
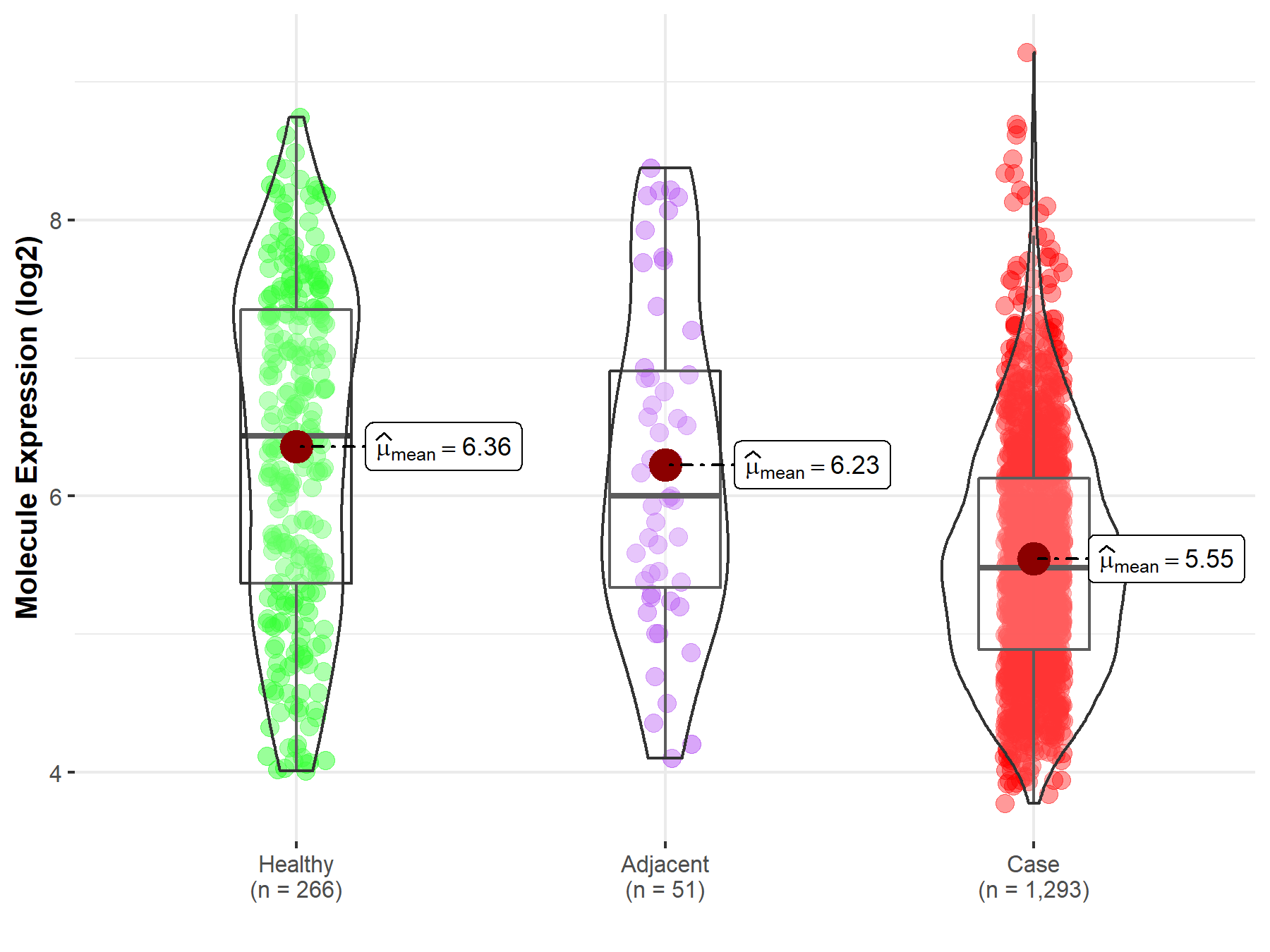
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.36E-03; Fold-change: -1.83E+00; Z-score: -2.11E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.40E-03; Fold-change: -9.62E-01; Z-score: -1.06E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
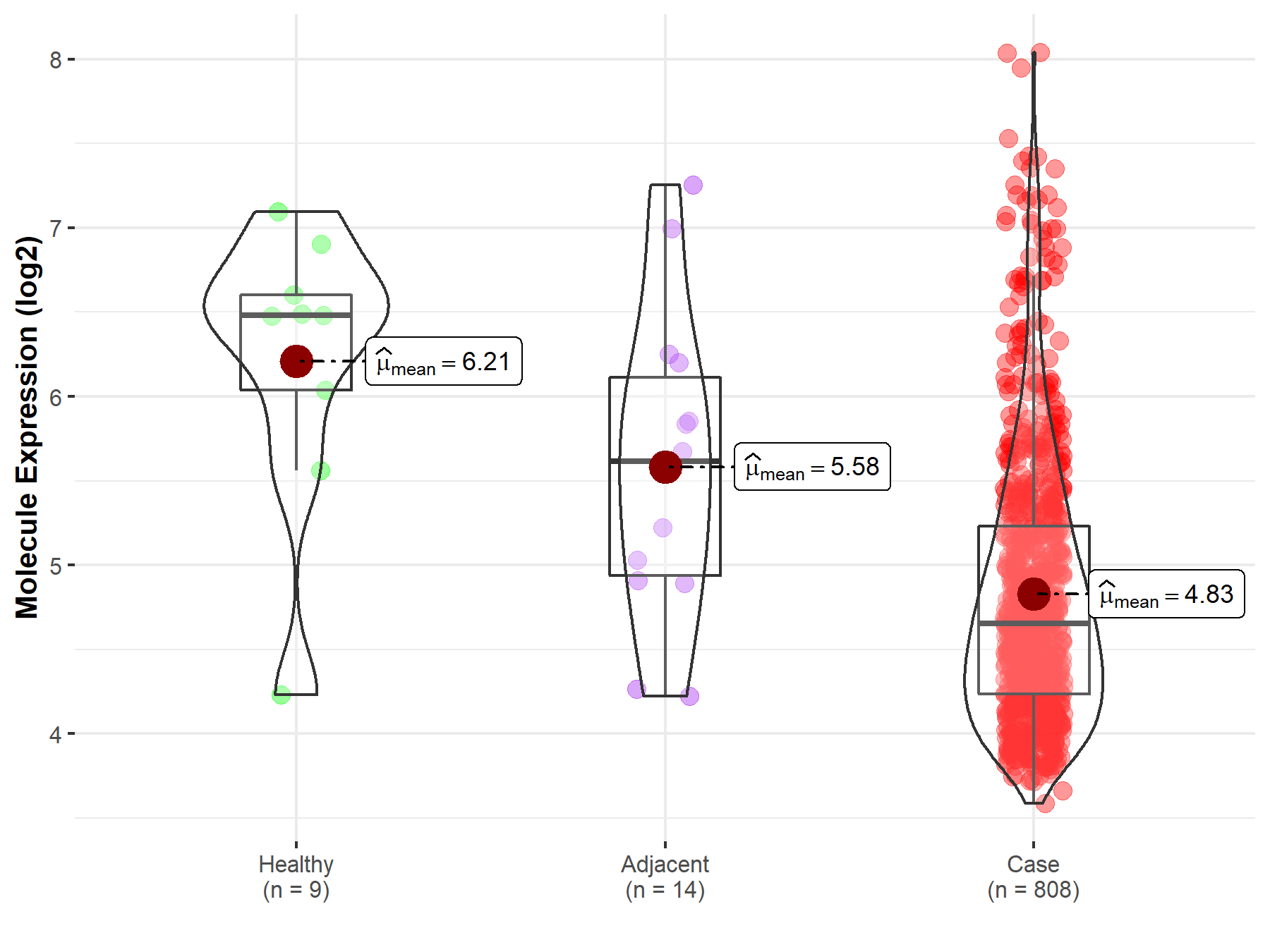
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
