Molecule Information
General Information of the Molecule (ID: Mol00186)
| Name |
TNF receptor-associated factor 6 (TRAF6)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
E3 ubiquitin-protein ligase TRAF6; Interleukin-1 signal transducer; RING finger protein 85; RING-type E3 ubiquitin transferase TRAF6; RNF85
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
TRAF6
|
||||
| Gene ID | |||||
| Location |
chr11:36483769-36510272[-]
|
||||
| Sequence |
MSLLNCENSCGSSQSESDCCVAMASSCSAVTKDDSVGGTASTGNLSSSFMEEIQGYDVEF
DPPLESKYECPICLMALREAVQTPCGHRFCKACIIKSIRDAGHKCPVDNEILLENQLFPD NFAKREILSLMVKCPNEGCLHKMELRHLEDHQAHCEFALMDCPQCQRPFQKFHINIHILK DCPRRQVSCDNCAASMAFEDKEIHDQNCPLANVICEYCNTILIREQMPNHYDLDCPTAPI PCTFSTFGCHEKMQRNHLARHLQENTQSHMRMLAQAVHSLSVIPDSGYISEVRNFQETIH QLEGRLVRQDHQIRELTAKMETQSMYVSELKRTIRTLEDKVAEIEAQQCNGIYIWKIGNF GMHLKCQEEEKPVVIHSPGFYTGKPGYKLCMRLHLQLPTAQRCANYISLFVHTMQGEYDS HLPWPFQGTIRLTILDQSEAPVRQNHEEIMDAKPELLAFQRPTIPRNPKGFGYVTFMHLE ALRQRTFIKDDTLLVRCEVSTRFDMGSLRREGFQPRSTDAGV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
E3 ubiquitin ligase that, together with UBE2N and UBE2V1, mediates the synthesis of 'Lys-63'-linked-polyubiquitin chains conjugated to proteins, such as IKBKG, IRAK1, AKT1 and AKT2. Also mediates ubiquitination of free/unanchored polyubiquitin chain that leads to MAP3K7 activation. Leads to the activation of NF-kappa-B and JUN. Seems to also play a role in dendritic cells (DCs) maturation and/or activation. Represses c-Myb-mediated transactivation, in B-lymphocytes. Adapter protein that seems to play a role in signal transduction initiated via TNF receptor, IL-1 receptor and IL-17 receptor. Regulates osteoclast differentiation by mediating the activation of adapter protein complex 1 (AP-1) and NF-kappa-B, in response to RANK-L stimulation. Together with MAP3K8, mediates CD40 signals that activate ERK in B-cells and macrophages, and thus may play a role in the regulation of immunoglobulin production.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [1] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioma | |||
| The Studied Tissue | Brainstem tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.53E-01 Fold-change: -2.62E-02 Z-score: -2.33E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AKT/NF-kappaB signaling pathway | Inhibition | hsa05135 | |
| In Vitro Model | U251-MG cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR 146b 5p suppresses glioblastoma cell resistance to temozolomide through targeting TRAF6. Overexpression of miR 146b 5p or TRAF6 knockdown significantly decreased the level of p AkT and p p65. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pain [ICD-11: MG30.0] | [2] | |||
| Resistant Disease | Pain [ICD-11: MG30.0] | |||
| Resistant Drug | Dezocine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative reverse-transcription PCR assay | |||
| Mechanism Description | miR-124-3p alleviates the dezocine tolerance against pain by regulating TRAF6 in a rat model. miR-124-3p expression was highly downregulated in a dezocine-resistant model. miR-124-3p overexpression could alleviate dezocine tolerance in rats. TRAF6 expression was significantly upregulated in a dezocine-resistant model. miR-124-3p targeted TRAF6 and TRAF6 was negatively modulated by miR-124-3p. In addition, overexpression of TRAF6 could reverse the inhibitory effects of miR-124-3p on dezocine tolerance. Overexpression of miR-124-3p alleviates dezocine tolerance against pain via regulating TRAF6 in a rat model, providing a possible solution to address dezocine tolerance in clinical. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [3] | |||
| Sensitive Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Sensitive Drug | Calycosin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| TRAF6-related signaling pathway | Regulation | N.A. | ||
| In Vitro Model | CNE2 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6889 |
| C666-1 cells | Throat | Homo sapiens (Human) | CVCL_7949 | |
| CNE1 cells | Throat | Homo sapiens (Human) | CVCL_6888 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; BrdU assay | |||
| Mechanism Description | Calycosin inhibits nasopharyngeal carcinoma cells by downregulating EWSAT1 expression to regulate the TRAF6-related pathways. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.88E-04; Fold-change: 1.00E-01; Z-score: 3.74E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.53E-01; Fold-change: -2.33E-01; Z-score: -3.01E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-03; Fold-change: 8.42E-01; Z-score: 1.47E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.62E-04; Fold-change: 1.77E-01; Z-score: 1.59E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
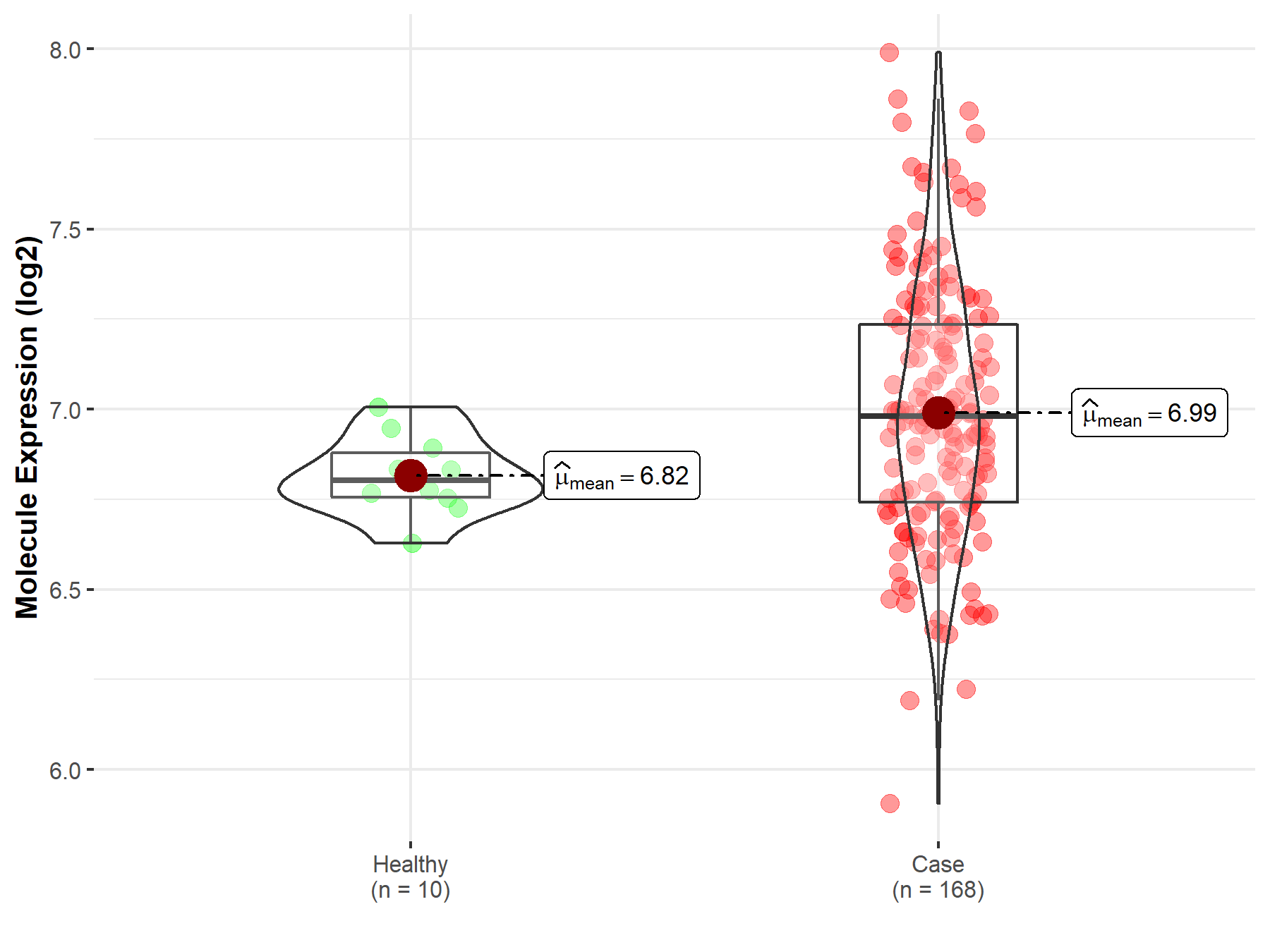
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
