Molecule Information
General Information of the Molecule (ID: Mol00128)
| Name |
Neurogenic locus notch homolog protein 1 (NOTCH1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Notch 1; hN1; Translocation-associated notch protein TAN-1; NEXT; NICD; TAN1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
NOTCH1
|
||||
| Gene ID | |||||
| Location |
chr9:136494433-136546048[-]
|
||||
| Sequence |
MPPLLAPLLCLALLPALAARGPRCSQPGETCLNGGKCEAANGTEACVCGGAFVGPRCQDP
NPCLSTPCKNAGTCHVVDRRGVADYACSCALGFSGPLCLTPLDNACLTNPCRNGGTCDLL TLTEYKCRCPPGWSGKSCQQADPCASNPCANGGQCLPFEASYICHCPPSFHGPTCRQDVN ECGQKPGLCRHGGTCHNEVGSYRCVCRATHTGPNCERPYVPCSPSPCQNGGTCRPTGDVT HECACLPGFTGQNCEENIDDCPGNNCKNGGACVDGVNTYNCRCPPEWTGQYCTEDVDECQ LMPNACQNGGTCHNTHGGYNCVCVNGWTGEDCSENIDDCASAACFHGATCHDRVASFYCE CPHGRTGLLCHLNDACISNPCNEGSNCDTNPVNGKAICTCPSGYTGPACSQDVDECSLGA NPCEHAGKCINTLGSFECQCLQGYTGPRCEIDVNECVSNPCQNDATCLDQIGEFQCICMP GYEGVHCEVNTDECASSPCLHNGRCLDKINEFQCECPTGFTGHLCQYDVDECASTPCKNG AKCLDGPNTYTCVCTEGYTGTHCEVDIDECDPDPCHYGSCKDGVATFTCLCRPGYTGHHC ETNINECSSQPCRHGGTCQDRDNAYLCFCLKGTTGPNCEINLDDCASSPCDSGTCLDKID GYECACEPGYTGSMCNINIDECAGNPCHNGGTCEDGINGFTCRCPEGYHDPTCLSEVNEC NSNPCVHGACRDSLNGYKCDCDPGWSGTNCDINNNECESNPCVNGGTCKDMTSGYVCTCR EGFSGPNCQTNINECASNPCLNQGTCIDDVAGYKCNCLLPYTGATCEVVLAPCAPSPCRN GGECRQSEDYESFSCVCPTGWQGQTCEVDINECVLSPCRHGASCQNTHGGYRCHCQAGYS GRNCETDIDDCRPNPCHNGGSCTDGINTAFCDCLPGFRGTFCEEDINECASDPCRNGANC TDCVDSYTCTCPAGFSGIHCENNTPDCTESSCFNGGTCVDGINSFTCLCPPGFTGSYCQH DVNECDSQPCLHGGTCQDGCGSYRCTCPQGYTGPNCQNLVHWCDSSPCKNGGKCWQTHTQ YRCECPSGWTGLYCDVPSVSCEVAAQRQGVDVARLCQHGGLCVDAGNTHHCRCQAGYTGS YCEDLVDECSPSPCQNGATCTDYLGGYSCKCVAGYHGVNCSEEIDECLSHPCQNGGTCLD LPNTYKCSCPRGTQGVHCEINVDDCNPPVDPVSRSPKCFNNGTCVDQVGGYSCTCPPGFV GERCEGDVNECLSNPCDARGTQNCVQRVNDFHCECRAGHTGRRCESVINGCKGKPCKNGG TCAVASNTARGFICKCPAGFEGATCENDARTCGSLRCLNGGTCISGPRSPTCLCLGPFTG PECQFPASSPCLGGNPCYNQGTCEPTSESPFYRCLCPAKFNGLLCHILDYSFGGGAGRDI PPPLIEEACELPECQEDAGNKVCSLQCNNHACGWDGGDCSLNFNDPWKNCTQSLQCWKYF SDGHCDSQCNSAGCLFDGFDCQRAEGQCNPLYDQYCKDHFSDGHCDQGCNSAECEWDGLD CAEHVPERLAAGTLVVVVLMPPEQLRNSSFHFLRELSRVLHTNVVFKRDAHGQQMIFPYY GREEELRKHPIKRAAEGWAAPDALLGQVKASLLPGGSEGGRRRRELDPMDVRGSIVYLEI DNRQCVQASSQCFQSATDVAAFLGALASLGSLNIPYKIEAVQSETVEPPPPAQLHFMYVA AAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNA SDGALMDDNQNEWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAP TPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSEEEEDAPAVISDFIYQGASL HNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQIL IRNRATDLDARMHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNN VDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLLDHFANRDITDHMDRLPRDI AQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRK PSSKGLACGSKEAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLP SPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGGGRLAFETGPPRLSHLPVAS GTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAP SLQHGMVGPLHSSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPA NIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQADVQPLGPSSLAVHTILPQE SPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQW SSSSPHSNVSDWSEGVSSPPTSMQSQIARIPEAFK Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A. Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
12 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [1] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.58E-10 Fold-change: -1.42E-01 Z-score: -6.55E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Notch signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-8 dye assay; Flow cytometry assay | |||
| Mechanism Description | miR-449a was involved in cisplatin resistance and the overexpression of miR449a increased cisplatin sensitivity mainly through inhibiting proliferation and promoting apoptosis and the direct downregulating the expression of NOTCH1. | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [7] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Notch1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| HFOB cells | Bone | Homo sapiens (Human) | CVCL_3708 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Transwell assay | |||
| Mechanism Description | miR-92a inhibited cell growth, migration, and enhanced cisplatin sensitivity of OS cell by downregulating Notch1. | |||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Notch1 signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating LncRNA Ak022798 expression. First, we found that Notch 1 was highly expressed in the cisplatin-resistant gastric cancer cell lines SGC7901/DDP and BGC823/DDP cells. Furthermore, we used siRNA to interfere with LncRNA Ak022798 expression, and found that the expression of MRP1 and P-glycoprotein decreased significantly in SGC7901/DDP and BGC823/DDP cells, and their apoptosis as well as the expressions of caspase 3 and caspase 8 obviously increased. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.11E-01 Fold-change: 2.74E-02 Z-score: 4.27E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch1 signaling pathway | Activation | hsa04330 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating LncRNA Ak022798 expression. First, we found that Notch 1 was highly expressed in the cisplatin-resistant gastric cancer cell lines SGC7901/DDP and BGC823/DDP cells. Furthermore, we used siRNA to interfere with LncRNA Ak022798 expression, and found that the expression of MRP1 and P-glycoprotein decreased significantly in SGC7901/DDP and BGC823/DDP cells, and their apoptosis as well as the expressions of caspase 3 and caspase 8 obviously increased. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [2] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colorectal cancer [ICD-11: 2B91] | |||
| The Specified Disease | Colorectal cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.67E-06 Fold-change: -9.64E-02 Z-score: -4.72E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| IGF-1R/AKT/S6 signaling pathway | Inhibition | hsa05225 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HCT-8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Ectopic expression of miR-139-5p sensitized CRC cells to 5-FU by increasing 5-FU-induced apoptosis. In addition, miR-139-5p inhibited the expression of the miR-139-5p target gene NOTCH-1 and its downstream molecules MRP-1 and BCL-2, two key MDR-associated genes. Furthermore, silencing NOTCH-1 expression promoted the chemotherapeutic effects of 5-FU, and up-regulation of NOTCH-1 abrogated miR-139-5p-mediated sensitization to 5-FU in LoVo and HCT-116 cells. | |||
| Disease Class: Colorectal carcinoma [ICD-11: 2B91.3] | [3] | |||
| Sensitive Disease | Colorectal carcinoma [ICD-11: 2B91.3] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colorectal cancer [ICD-11: 2B91] | |||
| The Specified Disease | Colorectal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.67E-06 Fold-change: -9.64E-02 Z-score: -4.72E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | miR139-5p reverses CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal carcinoma [ICD-11: 2B91.3] | [3] | |||
| Sensitive Disease | Colorectal carcinoma [ICD-11: 2B91.3] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colorectal cancer [ICD-11: 2B91] | |||
| The Specified Disease | Colorectal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.67E-06 Fold-change: -9.64E-02 Z-score: -4.72E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | miR139-5p reverses CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal carcinoma [ICD-11: 2B91.3] | [3] | |||
| Sensitive Disease | Colorectal carcinoma [ICD-11: 2B91.3] | |||
| Sensitive Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colorectal cancer [ICD-11: 2B91] | |||
| The Specified Disease | Colorectal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.67E-06 Fold-change: -9.64E-02 Z-score: -4.72E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | miR139-5p reverses CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.01E-13 Fold-change: -3.21E-02 Z-score: -7.63E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Notch signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-34a expression modulated breast cancer cells response to ADR by targeting Notch1 and Notch signaling pathway. | |||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [6] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.89E-08 Fold-change: -6.17E-02 Z-score: -6.03E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Notch1/HES1-PTEN/AKT signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Caspase-3 Activity kit assay | |||
| Mechanism Description | miR-760 inhibits Dox-resistance in HCC cells through inhibiting Notch1 and promoting PTEN expression. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a negatively regulates the expression of Notch1 at mRNA and protein levels, and overexpression of Mir-34A can increase the drug sensitivity of breast cancer cells to ADR. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [10] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR34a/Notch1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay | |||
| Mechanism Description | Primary and mature miR34a were suppressed by treatment with p53 RNAi or the dominant-negative p53 mutant in MCF7 cells. Ectopic miR34a expression reduced cancer stem cell properties and increased sensitivity to doxorubicin treatment by directly targeting NOTCH1. Furthermore, tumors from nude mice treated with miR34a were significantly smaller compared with those of mice treated with control lentivirus. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [8] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Notch signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Transwell assay | |||
| Mechanism Description | miR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.V1676A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.S1689P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.V1599M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [12] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| Notch1 signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-34a Attenuates Paclitaxel Resistance in Prostate Cancer Cells via Direct Suppression of JAG1/Notch1 Axis. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [13] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| Notch1 signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a negatively regulated cell proliferation, migration, and invasion and breast cancer stem cell propagation by downregulating Notch1. The expression of miR-34a was negatively correlated with tumor stages, metastasis, and Notch1 expression in breast cancer tissues. Furthermore, overexpression of miR-34a increased chemosensitivity of breast cancer cells to paclitaxel (PTX) by downregulating the Notch1 pathway. Mammosphere formation and expression of the stemness factor ALDH1 were also reduced in the cells treated with miR-34a and PTX compared to those treated with PTX alone. miR-34a inhibited breast cancer stemness and increased the chemosensitivity to PTX partially by downregulating the Notch1 pathway, suggesting that miR-34a/Notch1 play an important role in regulating breast cancer stem cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.V1676A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.V1599M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.S1689P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [14] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Rituximab | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Activation | hsa04330 | |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Mutations in NOTCH1 result in increased stability of an activated intracellular NOTCH1 isoform, which confers cell survival and apoptosis resistance, in part by sustaining expression of the anti-apoptotic protein Mcl-1, and promoting the activity of the key translational regulator eIF4E. Compared with wild-type cases, NOTCH1-mutated cases have progressive disease and significantly shorter survival, and demonstrate resistance to the anti-CD20 monoclo.l antibody rituximab, a phenotype thought to be associated with the low CD20 levels and dysregulation of histone deacetylases(HDAC)-mediated epigenetic repression of CD20 expression observed in NOTCH1-mutated CLL. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.V1599M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.S1689P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.V1676A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.V1676A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.V1599M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [11] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.S1689P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Notch signaling pathway | Regulation | N.A. | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [15] | |||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | |||
| Sensitive Drug | WNT-974 | |||
| Molecule Alteration | Missense mutation | p.C478F (c.1433G>T) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| FaDu cells | Pharynx | Homo sapiens (Human) | CVCL_1218 | |
| HN30 cells | Nasopharyngeal | Homo sapiens (Human) | CVCL_5525 | |
| CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 | |
| SCC25 cells | Oral | Homo sapiens (Human) | CVCL_1682 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 | |
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | |
| UMSCC cells | Oral cavity | Homo sapiens (Human) | CVCL_7707 | |
| Hs840T cells | Pharynx | Homo sapiens (Human) | CVCL_0942 | |
| Detroit 562 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1171 | |
| A-253 cells | Salivary gland | Homo sapiens (Human) | CVCL_1060 | |
| SCC-4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 | |
| Hs840.T cells | N.A. | N.A. | N.A. | |
| In Vivo Model | Nude mouse(or nude rat) xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
TaqMan assay | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
Preclinical Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.A1707T (c.5119G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.A1552G (c.4655C>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.A1552V (c.4655C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.E1567K (c.4699G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.A1570G (c.4709C>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.V1575A (c.4724T>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.V1599M (c.4795G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.V1676I (c.5026G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.I1680S (c.5039T>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.R1683W (c.5047C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | MRK-003 | |||
| Molecule Alteration | Missense mutation | p.A1707T (c.5119G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 |
| TNsBC cells | Breast | Homo sapiens (Human) | N.A. | |
| T-ALL cells | Bone marrow | Homo sapiens (Human) | CVCL_1736 | |
| HCC2218 cells | Breast | Homo sapiens (Human) | CVCL_1263 | |
| HCC1187 cells | Breast | Homo sapiens (Human) | CVCL_1247 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell Titer-Glo luminescent assay; Luciferase assay | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.11E-01; Fold-change: 2.85E-01; Z-score: 6.18E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.01E-08; Fold-change: 4.42E-01; Z-score: 2.22E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
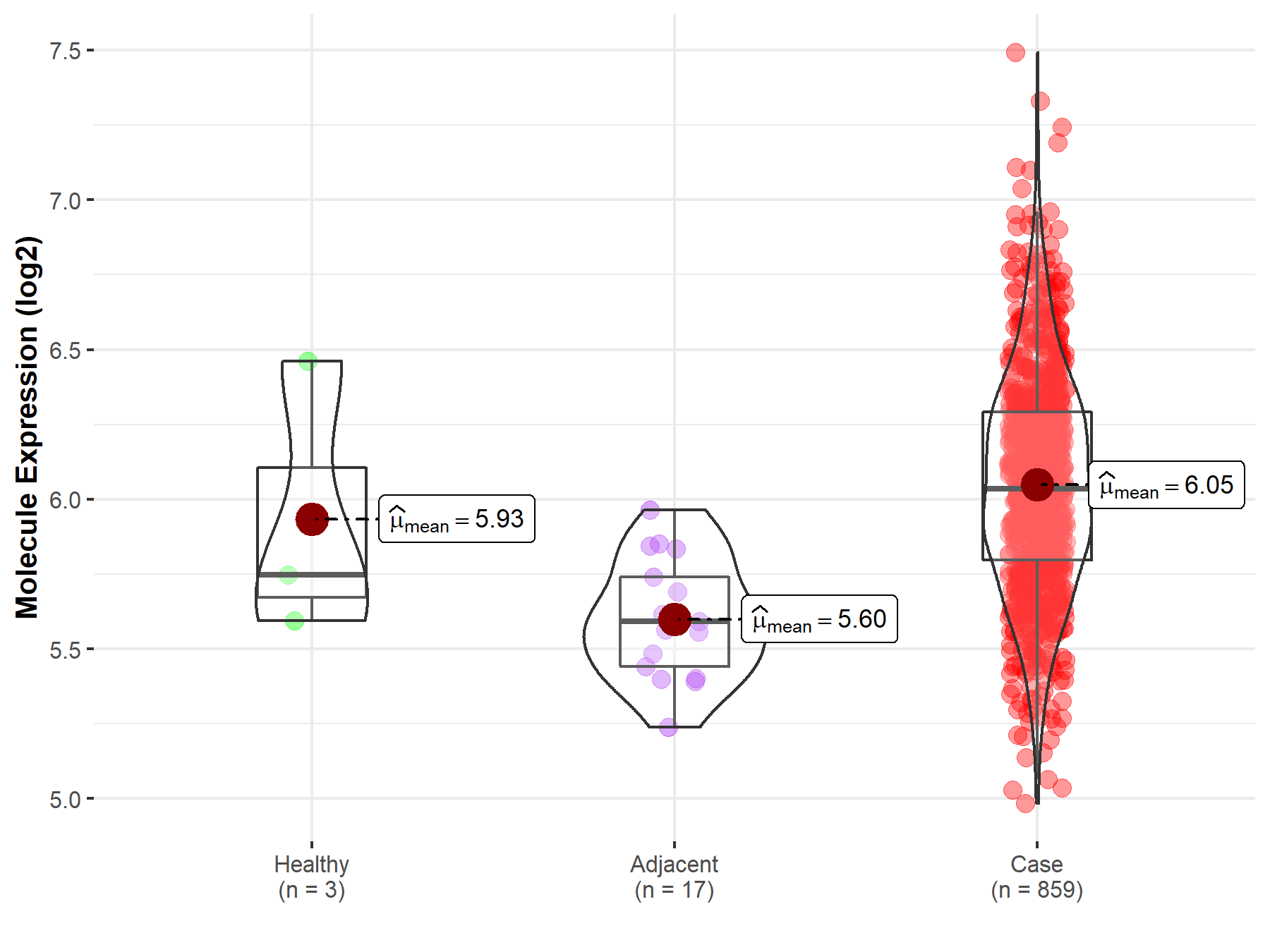
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.89E-08; Fold-change: -2.12E-01; Z-score: -9.02E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.45E-02; Fold-change: -5.94E-02; Z-score: -3.00E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 2.72E-01; Fold-change: 3.38E-02; Z-score: 1.01E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.01E-13; Fold-change: -1.70E-01; Z-score: -6.48E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.59E-01; Fold-change: -8.78E-02; Z-score: -2.31E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
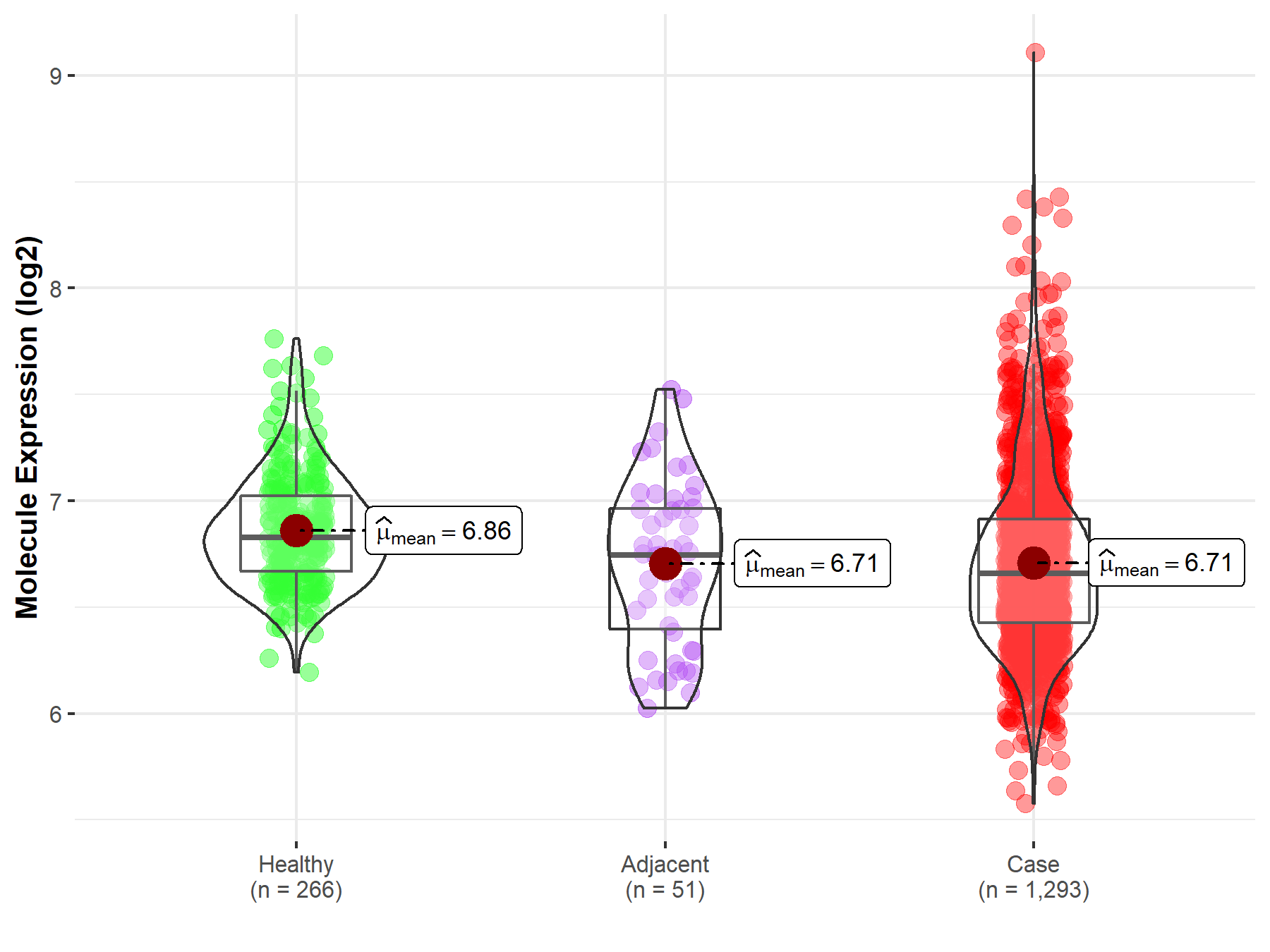
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.10E-01; Fold-change: -8.02E-03; Z-score: -2.14E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.59E-03; Fold-change: 2.54E-01; Z-score: 9.10E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
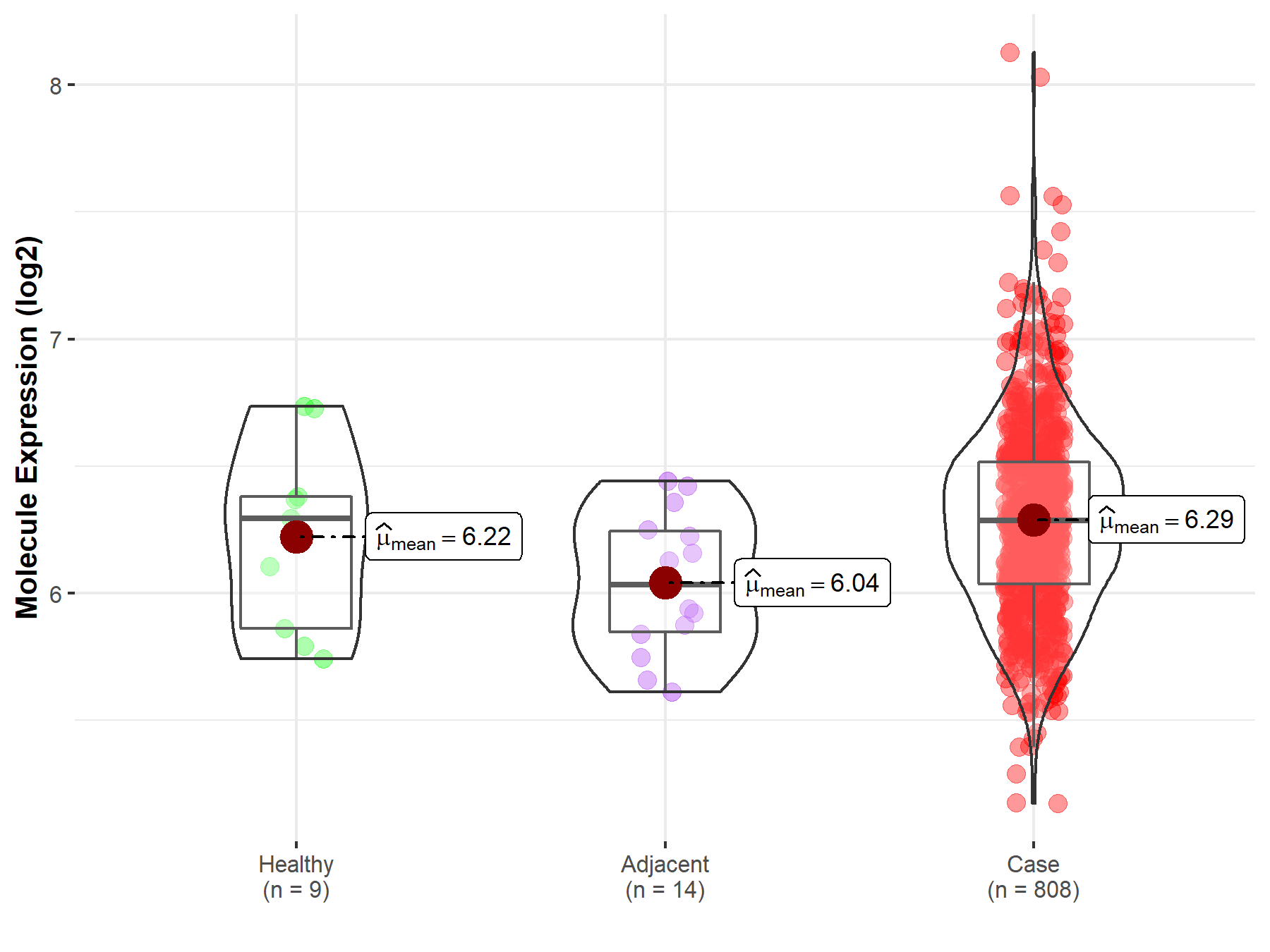
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.53E-01; Fold-change: 1.83E-01; Z-score: 4.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
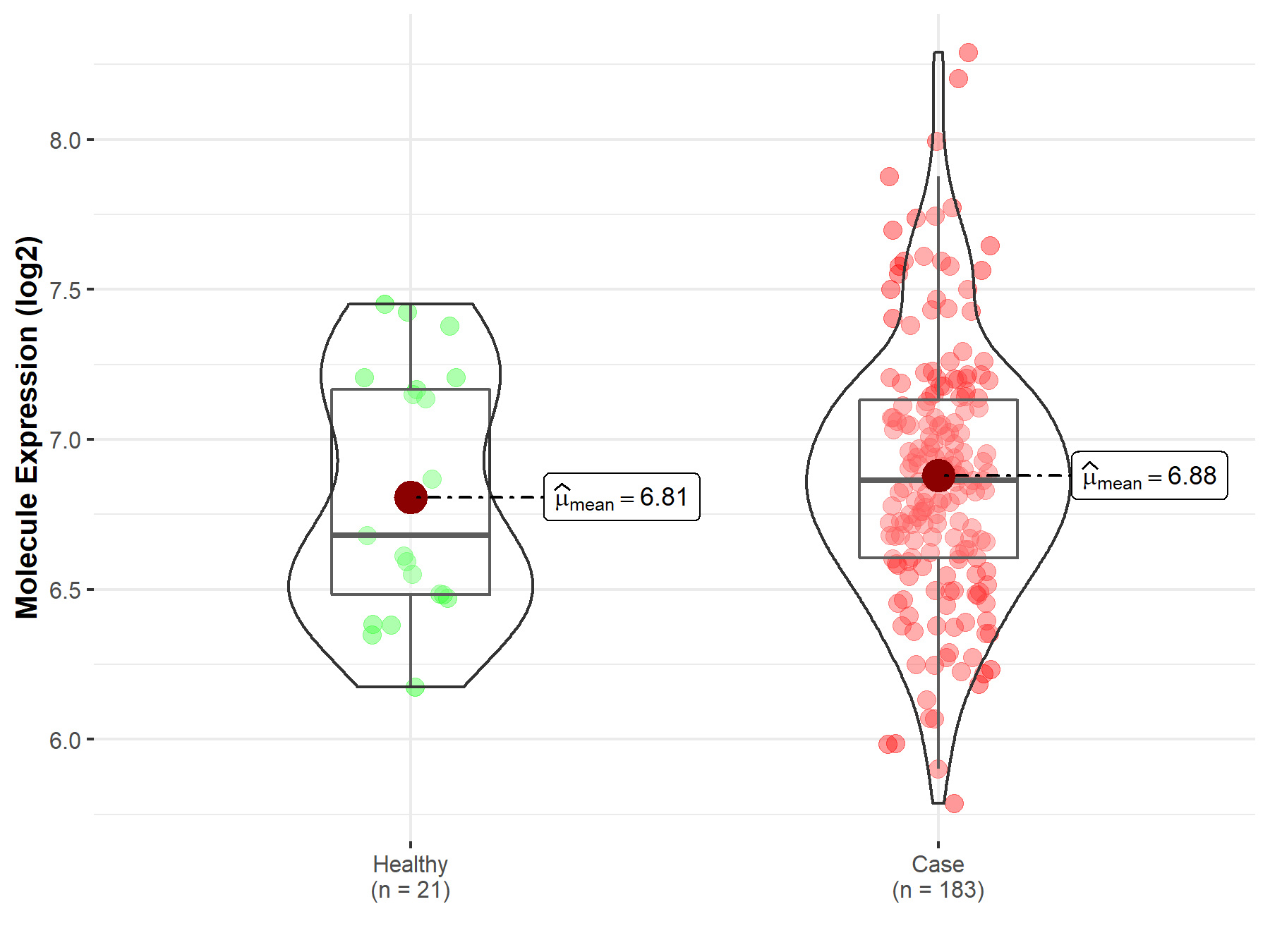
|
Click to View the Clearer Original Diagram |
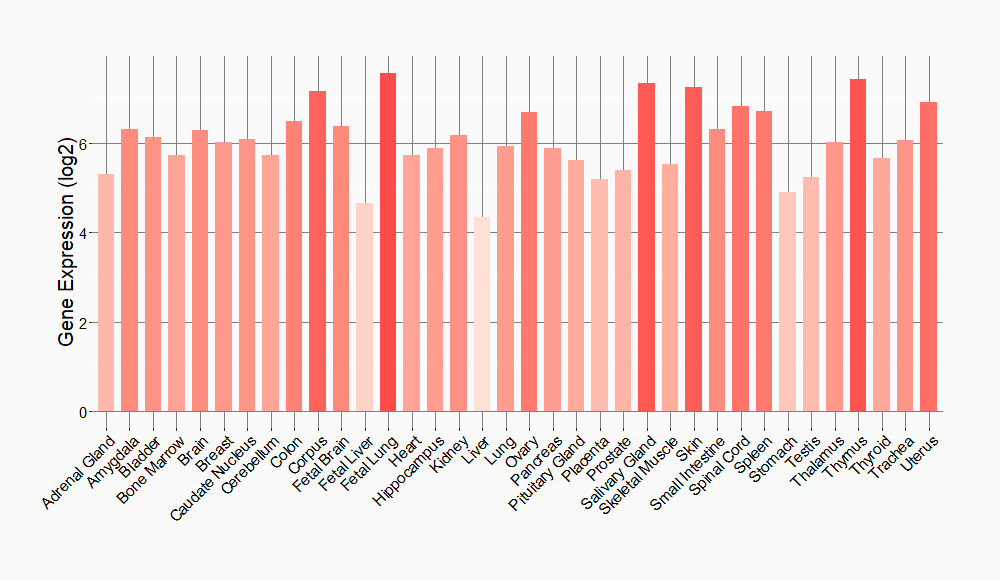
Tissue-specific Molecule Abundances in Healthy Individuals

|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
