Molecule Information
General Information of the Molecule (ID: Mol00036)
| Name |
Cadherin-1 (CDH1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Molecule Type |
Protein
|
||||
| Gene Name |
CDH1
|
||||
| Gene ID | |||||
| Location |
chr16:68737292-68835537[+]
|
||||
| Sequence |
MGPWSRSLSALLLLLQVSSWLCQEPEPCHPGFDAESYTFTVPRRHLERGRVLGRVNFEDC
TGRQRTAYFSLDTRFKVGTDGVITVKRPLRFHNPQIHFLVYAWDSTYRKFSTKVTLNTVG HHHRPPPHQASVSGIQAELLTFPNSSPGLRRQKRDWVIPPISCPENEKGPFPKNLVQIKS NKDKEGKVFYSITGQGADTPPVGVFIIERETGWLKVTEPLDRERIATYTLFSHAVSSNGN AVEDPMEILITVTDQNDNKPEFTQEVFKGSVMEGALPGTSVMEVTATDADDDVNTYNAAI AYTILSQDPELPDKNMFTINRNTGVISVVTTGLDRESFPTYTLVVQAADLQGEGLSTTAT AVITVTDTNDNPPIFNPTTYKGQVPENEANVVITTLKVTDADAPNTPAWEAVYTILNDDG GQFVVTTNPVNNDGILKTAKGLDFEAKQQYILHVAVTNVVPFEVSLTTSTATVTVDVLDV NEAPIFVPPEKRVEVSEDFGVGQEITSYTAQEPDTFMEQKITYRIWRDTANWLEINPDTG AISTRAELDREDFEHVKNSTYTALIIATDNGSPVATGTGTLLLILSDVNDNAPIPEPRTI FFCERNPKPQVINIIDADLPPNTSPFTAELTHGASANWTIQYNDPTQESIILKPKMALEV GDYKINLKLMDNQNKDQVTTLEVSVCDCEGAAGVCRKAQPVEAGLQIPAILGILGGILAL LILILLLLLFLRRRAVVKEPLLPPEDDTRDNVYYYDEEGGGEEDQDFDLSQLHRGLDARP EVTRNDVAPTLMSVPRYLPRPANPDEIGNFIDENLKAADTDPTAPPYDSLLVFDYEGSGS EAASLSSLNSSESDKDQDYDYLNEWGNRFKKLADMYGGGEDD Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Cadherins are calcium-dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types. CDH1 is involved in mechanisms regulating cell-cell adhesions, mobility and proliferation of epithelial cells. Has a potent invasive suppressor role. It is a ligand for integrin alpha-E/beta-7.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
11 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [1] | |||
| Resistant Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Cervical cancer [ICD-11: 2C77] | |||
| The Specified Disease | Cervical cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.15E-02 Fold-change: -8.66E-02 Z-score: -2.06E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | Siha cells | Cervix uteri | Homo sapiens (Human) | CVCL_0032 |
| Caski cells | Uterus | Homo sapiens (Human) | CVCL_1100 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Paclitaxel transiently induced up-regulation of miR-375 expression, proliferation inhibition, transition from epithelial to mesenchymal phenotype, and consequently impaired paclitaxel sensitivity. Forced over-expression of miR-375 may suppress Ecadherin expression by a directly targeting pathway, which led to paclitaxel resistance. Contrarily, re-expression of Ecadherin partly reversed epithelial-mesenchymal transition phenotype and miR-375 induced paclitaxel-resistance. Our findings suggest that paclitaxel-induced miR-375 over-expression facilitates epithelial-mesenchymal transition process via directly targeting Ecadherin, proliferation inhibition, and consequently results in chemo-resistance in cervical cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.17E-05 Fold-change: 1.76E-01 Z-score: 4.06E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Wnt signaling pathway | Inhibition | hsa04310 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of H19 by siRNA transfection can significantly reduce the expression of N-cadherin, as well as increase E-cadherin and vimentin level, which improved tamoxifen sensitivity in tamoxifen-resistant breast cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [3] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colorectal cancer [ICD-11: 2B91] | |||
| The Specified Disease | Colorectal cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.45E-01 Fold-change: 6.54E-03 Z-score: 1.95E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V/ PI staining; Caspase-3 activity assay | |||
| Mechanism Description | Levels of PTEN and E-cadherin were reduced by knockdown of miR200c in HCT-116 cells, PTEN inactivate the AkT signaling pathway, and E-cadherin is one of the major downstream regulators of miRNA-200c contributing to EMT, which is also important to inhibit tumor invasion and proliferation as well as to induce cell apoptosis. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic carcinoma [ICD-11: 2C10.2] | [4] | |||
| Sensitive Disease | Pancreatic carcinoma [ICD-11: 2C10.2] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.75E-02 Fold-change: 1.34E-01 Z-score: 2.27E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Colorimetric methylene blue assay; Flow cytometry assay | |||
| Mechanism Description | Forced expression of miR-200b induces CDH1 expression and promotes gemcitabine sensitivity in Capan-2 and Panc-1 cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.24E-15 Fold-change: 1.16E-01 Z-score: 8.37E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [6] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| N-Myc/ miR421 /ATM signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| HGC27 cells | Gastric | Homo sapiens (Human) | CVCL_1279 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| SNU-16 cells | Gastric | Homo sapiens (Human) | CVCL_0076 | |
| Experiment for Molecule Alteration |
Western blot analysis; Flow cytometric assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Overexpression of miR-421 promoted metastasis, inhibited apoptosis, and induced cisplatin resistance in gastric cancer by targeting E-cadherin and caspase-3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [7] | |||
| Sensitive Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 |
| SCC25 cells | Oral | Homo sapiens (Human) | CVCL_1682 | |
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | |
| SCC15 cells | Tongue | Homo sapiens (Human) | CVCL_1681 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | HULC-depleted cells showed decreased expression of vimentin and N-cadherin and increased expression of E-cadherin, which shows that HULC participates in the EMT process and affects the expression levels of proteins that are crucial for cell proliferation and invasion. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.V345A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.A348V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.R90Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Missense mutation | p.R74Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [9] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| FHC cells | Colon | Homo sapiens (Human) | CVCL_3688 | |
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Boyden chambers cell migration and invasion assays | |||
| Mechanism Description | MALAT1 tethers EZH2 to CDH1 promoter and suppresses miR218 during oxaliplatin treatment, which finally promotes colorectal cancer cell EMT, metastasis, and chemoresistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.A348V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.V345A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.R90Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Pertuzumab | |||
| Molecule Alteration | Missense mutation | p.R74Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.R90Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.A348V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.V345A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.R74Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.A348V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.V345A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.R90Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [8] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab emtansine | |||
| Molecule Alteration | Missense mutation | p.R74Q |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | |||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Oral tissue | |
| The Specified Disease | Oral squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.21E-01; Fold-change: 6.52E-01; Z-score: 7.44E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.43E-02; Fold-change: -6.28E-01; Z-score: -4.67E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
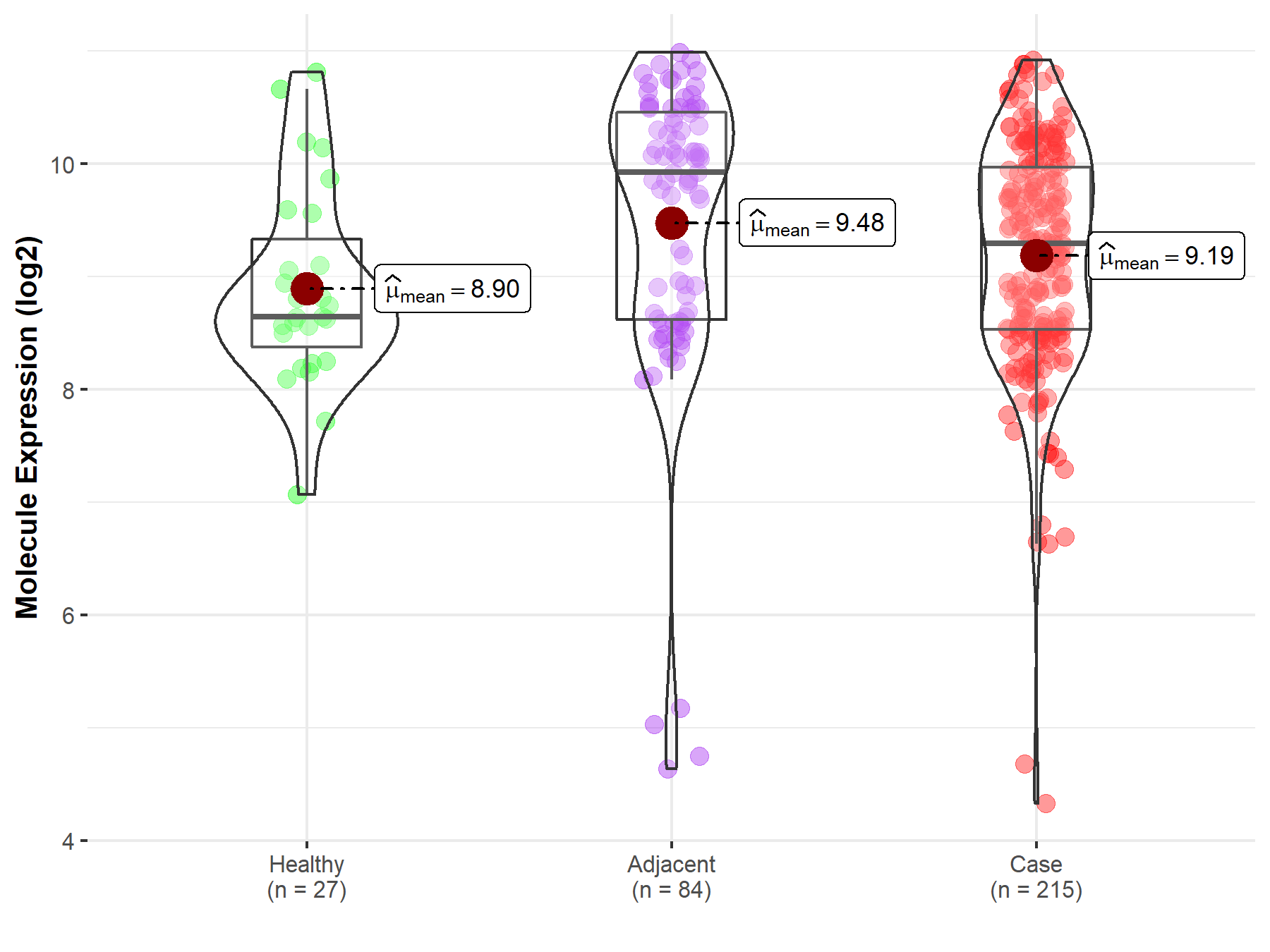
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.99E-01; Fold-change: -1.32E-02; Z-score: -7.56E-03 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.81E-03; Fold-change: 1.20E-01; Z-score: 3.61E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.75E-02; Fold-change: 7.20E-01; Z-score: 5.60E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.61E-01; Fold-change: -3.56E-01; Z-score: -2.76E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
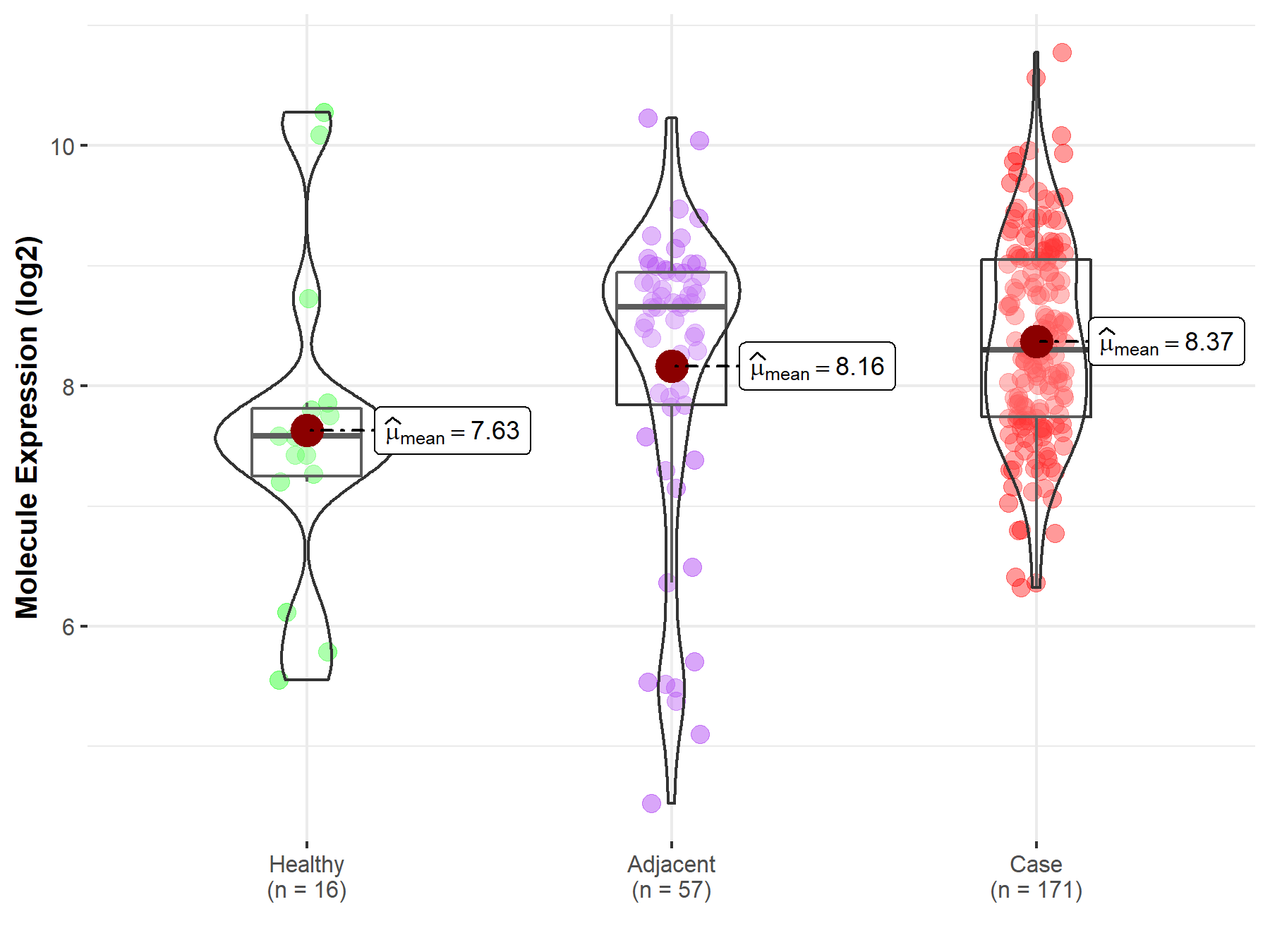
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.24E-15; Fold-change: 5.22E-01; Z-score: 4.26E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.24E-04; Fold-change: 4.65E-01; Z-score: 3.98E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.48E-01; Fold-change: -1.65E-01; Z-score: -6.45E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

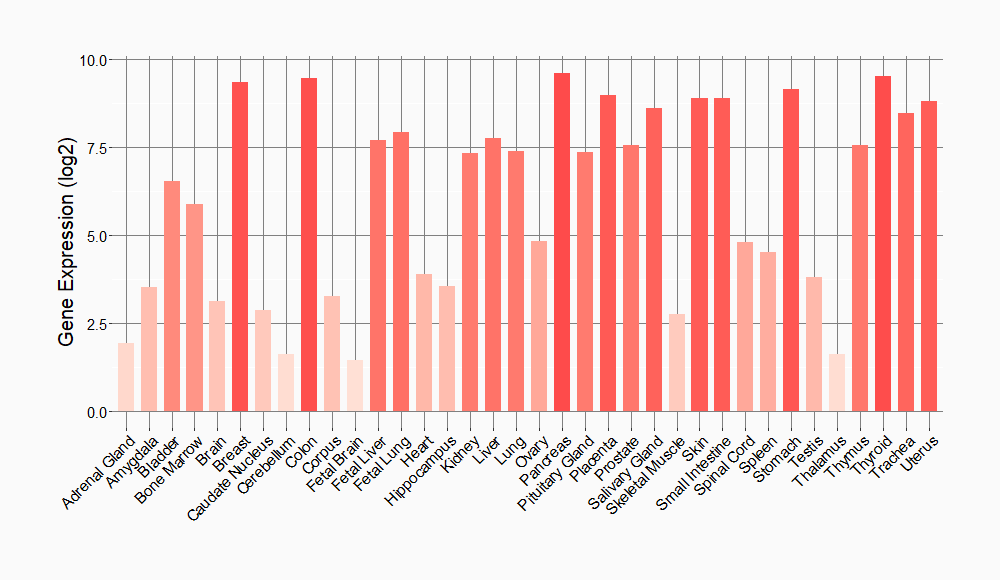
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
