Molecule Information
General Information of the Molecule (ID: Mol00024)
| Name |
Bcl-2-like protein 11 (BCL2L11)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Bcl2-L-11; Bcl2-interacting mediator of cell death; BIM
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
BCL2L11
|
||||
| Gene ID | |||||
| Location |
chr2:111119378-111168445[+]
|
||||
| Sequence |
MAKQPSDVSSECDREGRQLQPAERPPQLRPGAPTSLQTEPQGNPEGNHGGEGDSCPHGSP
QGPLAPPASPGPFATRSPLFIFMRRSSLLSRSSSGYFSFDTDRSPAPMSCDKSTQTPSPP CQAFNHYLSAMASMRQAEPADMRPEIWIAQELRRIGDEFNAYYARRVFLNNYQAAEDHPR MVILRLLRYIVRLVWRMH Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Induces apoptosis and anoikis. Isoform BimL is more potent than isoform BimEL. Isoform Bim-alpha1, isoform Bim-alpha2 and isoform Bim-alpha3 induce apoptosis, although less potent than isoform BimEL, isoform BimL and isoform BimS. Isoform Bim-gamma induces apoptosis. Isoform Bim-alpha3 induces apoptosis possibly through a caspase-mediated pathway. Isoform BimAC and isoform BimABC lack the ability to induce apoptosis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [1] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.55E-01 Fold-change: -6.57E-02 Z-score: -1.43E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR138/BIM signaling pathway | Regulation | N.A. | ||
| In Vitro Model | LN229 cells | Brain | Homo sapiens (Human) | CVCL_0393 |
| A172 cells | Brain | Homo sapiens (Human) | CVCL_0131 | |
| LN-18 cells | Brain | Homo sapiens (Human) | CVCL_0392 | |
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | |
| U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |
| LN308 cells | Brain | Homo sapiens (Human) | CVCL_0394 | |
| D247MG cells | Brain | Homo sapiens (Human) | CVCL_1153 | |
| LN-319 cells | Brain | Homo sapiens (Human) | CVCL_3958 | |
| LN-428 cells | Brain | Homo sapiens (Human) | CVCL_3959 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | Transient transfection of miR-138 mimics in glioma cells with low basal miR-138 expression increased glioma cell proliferation. Moreover, miR-138 overexpression increased TMZ resistance in long-term glioblastoma cell lines and glioma initiating cell cultures. The apoptosis regulator BIM was identified as a direct target of miR-138, and its silencing mediated the induced TMZ resistance phenotype. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pediatric intracranial nongerminomatous malignant germ cell tumors [ICD-11: 2A00.07] | [2] | |||
| Resistant Disease | Pediatric intracranial nongerminomatous malignant germ cell tumors [ICD-11: 2A00.07] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Pediatric intracranial nongerminomatous malignant germ cell tumors | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.55E-01 Fold-change: -6.57E-02 Z-score: -1.43E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Immunoblotting assay; Immunohistochemistry | |||
| Experiment for Drug Resistance |
MTT Assay | |||
| Mechanism Description | miR214-3p overexpression enhanced cisplatin resistance by downregulating the expression of its target, the apoptotic protein BCL2-like 11 (BCL2L11/BIM). | |||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [3] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Prostate cancer [ICD-11: 2C82] | |||
| The Specified Disease | Prostate cancer | |||
| The Studied Tissue | Prostate | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.20E-04 Fold-change: -1.26E-01 Z-score: -3.89E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Activation | hsa04010 | |
| Cell apoptosis | Inhibition | hsa04210 | ||
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-17-92 cluster plays a crucial role in cell growth of the DU145 prostate cancer cells due to regulation of cellular apoptosis-related and proliferation-related proteins, and causes chemo-resistance to cisplatin via activating AkT signaling together with upregulating ERCC1 also contributed to development of cisplatin-resistance. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [4] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-192 induced Cisplatin-resistance and inhibited cell apoptosis in lung cancer via negative targeting Bim expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Myeloid leukemia [ICD-11: 2A60.4] | [5] | |||
| Sensitive Disease | Myeloid leukemia [ICD-11: 2A60.4] | |||
| Sensitive Drug | Cytarabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | One of the predicted targets of miR-32 lies in the 3' untranslated region (UTR) of BCL2L11 gene, which encodes the pro-apoptotic protein Bim, miR-32 blockade is sufficient to elevate Bim expression and sensitize AML cells to chemotherapy-induced apoptosis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [6] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Bim signaling pathway | Activation | hsa05206 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [7] | |||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/BCL2 signaling pathway | Regulation | N.A. | |
| Cell apoptosis | Activation | hsa04210 | ||
| NF-kappaB signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | microRNA-184 modulates doxorubicin resistance in osteosarcoma cells by targeting BCL2L1 and enhancing the level of it. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [8] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| miR181b/Bim/MMP/caspase signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-181b level was significantly upregulated in patient serum and breast cancer cell lines compared with that in normal controls. The results of in vitro 3H thymidine incorporation and Transwell migration assay indicated that miR-181b overexpression markedly promoted the proliferation and metastasis of breast cancer cells. These data suggest that miR-181b is a tumor promoter in breast cancer. Furthermore, miR-181b expression was found to be upregulated in doxorubicin (DOX)-resistant T-47D cells (T-47D-R) compared with that in the parental T-47D cells, and upregulation of miR-181b expression decreased the anticancer effect of DOX in the T-47D cells. Mechanistic studies demonstrated that the Bim gene, an essential initiator of apoptosis, was inhibited by miR-181b overexpression. knockdown of miR-181b by its specific inhibitors significantly re-sensitized the T-47D-R cells to the cytotoxicity of DOX. miR-181b inhibitors increased the level of Bim in the T-47D-R cells, resulting in the loss of mitochondrial membrane potential (MMP) and the activation of caspases caused by DOX. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [9] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR10b/BIM signaling pathway | Activation | hsa05206 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| Experiment for Molecule Alteration |
Luciferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-10b directly inhibits pro-apoptotic BIM, and the overexpression of miR-10b confers chemoresistance in colorectal cancer cells to 5-FU. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [10] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Venetoclax | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | H1048 shp53 cells | Lung | Homo sapiens (Human) | N.A. |
| H211 shp53 cells | Lung | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot assay; qRT-PCR | |||
| Experiment for Drug Resistance |
Fluorescence-activated cell sorting assay; Cell viability assay | |||
| Mechanism Description | Down-Regulation of Onc-p53 Increases BIM Expression and Sensitizes to Venetoclax in SCLC-P Cells. Down-regulation of Onc-p53 increases the expression of a BH3-only pro-apoptotic BIM and sensitizes to venetoclax in SCLC-P cells | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.54E-132; Fold-change: 8.01E-01; Z-score: 2.09E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.09E-01; Fold-change: 6.11E-01; Z-score: 1.05E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.68E-01; Fold-change: 1.53E-01; Z-score: 2.72E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
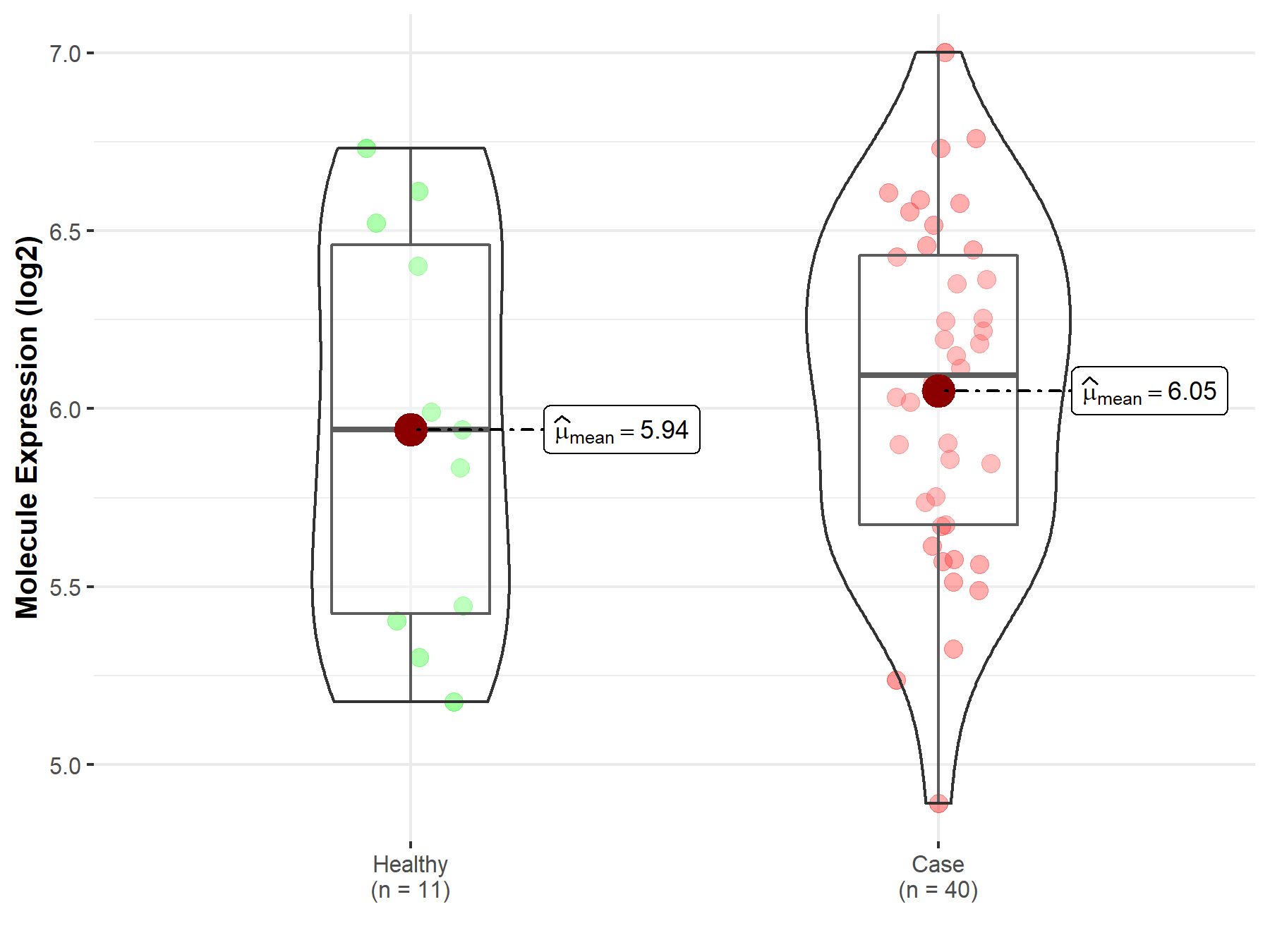
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.47E-04; Fold-change: 4.34E-01; Z-score: 1.82E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.05E-13; Fold-change: -3.53E-01; Z-score: -7.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
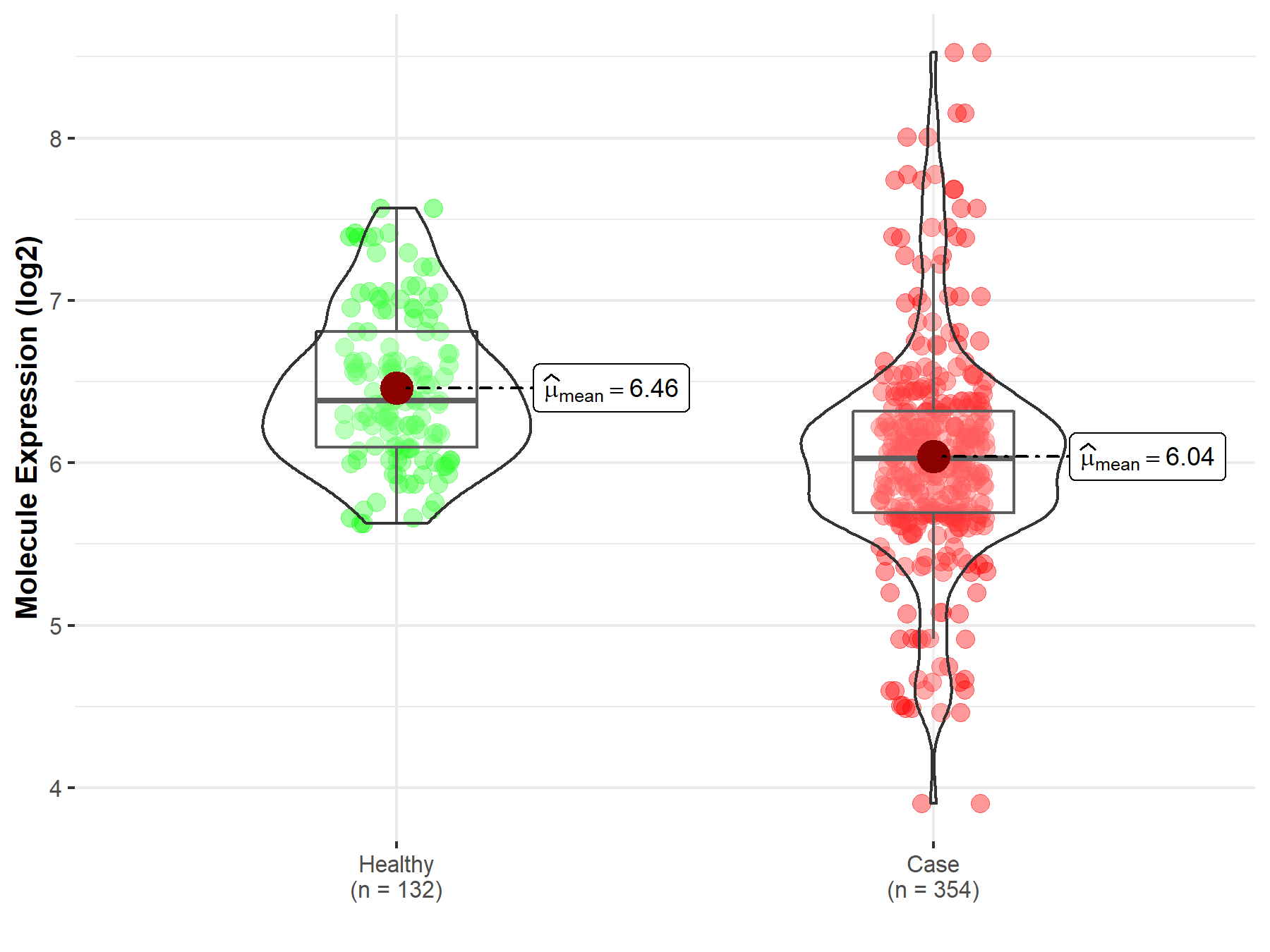
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.77E-37; Fold-change: 2.78E-01; Z-score: 1.02E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.81E-14; Fold-change: 2.03E-01; Z-score: 6.02E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.52E-03; Fold-change: -2.82E-03; Z-score: -8.18E-03 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.17E-01; Fold-change: 1.91E-02; Z-score: 4.14E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.20E-04; Fold-change: -9.04E-01; Z-score: -1.64E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
