Drug Information
Drug (ID: DG01641) and It's Reported Resistant Information
| Name |
Enasidenib
|
||||
|---|---|---|---|---|---|
| Synonyms |
Enasidenib; 1446502-11-9; AG-221; AG-221 (Enasidenib); CC-90007 Free Base; IDHIFA; UNII-3T1SS4E7AG; AG 221; 2-Methyl-1-(4-(6-(trifluoromethyl)pyridin-2-yl)-6-(2-(trifluoromethyl)pyridin-4-ylamino)-1,3,5-triazin-2-ylamino)propan-2-ol; CC-90007; 3T1SS4E7AG; 2-methyl-1-((4-(6-(trifluoromethyl)pyridin-2-yl)-6-((2-(trifluoromethyl)pyridin-4-yl)amino)-1,3,5-triazin-2-yl)amino)propan-2-ol; 2-Methyl-1-[(4-[6-(Trifluoromethyl)pyridin-2-Yl]-6-{[2-(Trifluoromethyl)pyridin-4-Yl]amino}-1,3,5-Triazin-2-Yl)amino]propan-2-Ol; 2-methyl-1-[[4-[6-(trifluoromethyl)pyridin-2-yl]-6-[[2-(trifluoromethyl)pyridin-4-yl]amino]-1,3,5-triazin-2-yl]amino]propan-2-ol; 2-Propanol, 2-methyl-1-[[4-[6-(trifluoromethyl)-2-pyridinyl]-6-[[2-(trifluoromethyl)-4-pyridinyl]amino]-1,3,5-triazin-2-yl]amino]-; 2-Propanol, 2-methyl-1-((4-(6-(trifluoromethyl)-2-pyridinyl)-6-((2-(trifluoromethyl)-4-pyridinyl)amino)-1,3,5-triazin-2-yl)amino)-; Enasidenib [INN]; enasidenibum; AG221; 2-methyl-1-({4-[6-(trifluoromethyl)pyridin-2-yl]-6-{[2-(trifluoromethyl)pyridin-4-yl]amino}-1,3,5-triazin-2-yl}amino)propan-2-ol; AG-221(Enasidenib); AG-221; Enasidenib; Enasidenib; AG-221; Enasidenib (USAN/INN); Enasidenib [USAN:INN]; GTPL8960; CHEMBL3989908; SCHEMBL15102202; EX-A654; CHEBI:145374; HMS3873D03; AMY38698; BCP16041; BDBM50503251; MFCD29472245; NSC788120; s8205; AKOS026750439; ZINC222731806; CCG-269476; CS-5017; DB13874; NSC-788120; SB19193; NCGC00479249-03; NCGC00479249-05; AC-31318; AS-75164; HY-18690; FT-0700204; D10901; A857662; J-690181; Q27077182; B0084-470859; AG-221; AG 221; AG221; CC-90007; CC 90007; CC90007; 69Q
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
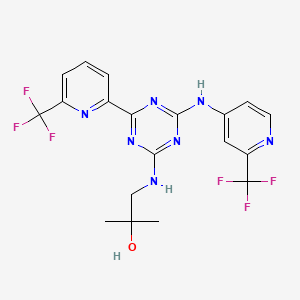
|
||||
| Target | Serine/threonine-protein kinase B-raf (BRAF) | BRAF_HUMAN | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
6
|
||||
| IsoSMILES |
CC(C)(CNC1=NC(=NC(=N1)C2=NC(=CC=C2)C(F)(F)F)NC3=CC(=NC=C3)C(F)(F)F)O
|
||||
| InChI |
InChI=1S/C19H17F6N7O/c1-17(2,33)9-27-15-30-14(11-4-3-5-12(29-11)18(20,21)22)31-16(32-15)28-10-6-7-26-13(8-10)19(23,24)25/h3-8,33H,9H2,1-2H3,(H2,26,27,28,30,31,32)
|
||||
| InChIKey |
DYLUUSLLRIQKOE-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [1] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172K (c.515G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
40
|
-
A
-
D
-
K
-
R
-
I
-
K
-
V
-
A
-
K
-
P
50
|
-
V
-
V
-
E
-
M
-
D
-
G
-
D
-
E
-
M
-
T
60
|
-
R
-
I
-
I
-
W
-
Q
-
F
-
I
-
K
-
E
-
K
70
|
-
L
-
I
-
L
-
P
-
H
-
V
-
D
-
I
-
Q
-
L
80
|
-
K
-
Y
-
F
-
D
-
L
-
G
-
L
-
P
-
N
-
R
90
|
-
D
-
Q
-
T
-
D
-
D
-
Q
-
V
-
T
-
I
-
D
100
|
-
S
-
A
-
L
-
A
-
T
-
Q
-
K
-
Y
-
S
-
V
110
|
-
A
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
120
|
-
E
-
A
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
130
|
-
K
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
140
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
150
|
-
E
-
P
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
160
|
-
L
-
V
-
P
-
G
-
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
S
K
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
-
A
-
T
-
D
-
F
-
V
-
A
-
D
-
R
-
A
190
|
-
G
-
T
-
F
-
K
-
M
-
V
-
F
-
T
-
P
-
K
200
|
-
D
-
G
-
S
-
G
-
V
-
K
-
E
-
W
-
E
-
V
210
|
-
Y
-
N
-
F
-
P
-
A
-
G
-
G
-
V
-
G
-
M
220
|
-
G
-
M
-
Y
-
N
-
T
-
D
-
E
-
S
-
I
-
S
230
|
-
G
-
F
-
A
-
H
-
S
-
C
-
F
-
Q
-
Y
-
A
240
|
-
I
-
Q
-
K
-
K
-
W
-
P
-
L
-
Y
-
M
-
S
250
|
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
A
-
Y
-
D
260
|
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
-
I
270
|
-
F
-
D
-
K
-
H
-
Y
-
K
-
T
-
D
-
F
-
D
280
|
-
K
-
N
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
-
L
290
|
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
V
-
L
-
K
300
|
-
S
-
S
-
G
-
G
-
F
-
V
-
W
-
A
-
C
-
K
310
|
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
-
I
320
|
-
L
-
A
-
Q
-
G
-
F
-
G
-
S
-
L
-
G
-
L
330
|
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
-
G
340
|
-
K
-
T
-
I
-
E
-
A
-
E
-
A
-
A
-
H
-
G
350
|
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
E
-
H
-
Q
360
|
-
K
-
G
-
R
-
P
-
T
-
S
-
T
-
N
-
P
-
I
370
|
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
-
L
380
|
-
E
-
H
-
R
-
G
-
K
-
L
-
D
-
G
-
N
-
Q
390
|
-
D
-
L
-
I
-
R
-
F
-
A
-
Q
-
M
-
L
-
E
400
|
-
K
-
V
-
C
-
V
-
E
-
T
-
V
-
E
-
S
-
G
410
|
-
A
-
M
-
T
-
K
-
D
-
L
-
A
-
G
-
C
-
I
420
|
-
H
-
G
-
L
-
S
-
N
-
V
-
K
-
L
-
N
-
E
430
|
-
H
-
F
-
L
-
N
-
T
-
T
-
D
-
F
-
L
-
D
440
|
-
T
-
I
-
K
-
S
-
N
-
L
-
D
-
R
-
A
-
L
450
|
-
G
-
R
-
Q
-
S
-
L
-
E
-
H
-
H
-
H
-
H
460
|
-
H
-
H
-
H
-
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |||||||||
| TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | ||||||||||
| TF-1a cells | Bone marrow | Homo sapiens (Human) | CVCL_3608 | ||||||||||
| IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | NSG mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Somatic gain-of-function mutations in isocitrate dehydrogenases (IDH) 1 and 2 are found in multiple hematologic and solid tumors, leading to accumulation of the oncometabolite (R)-2-hydroxyglutarate (2HG). 2HG competitively inhibits alpha-ketoglutarate-dependent dioxygenases, including histone demethylases and methylcytosine dioxygenases of the TET family, causing epigenetic dysregulation and a block in cellular differentiation. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [1] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140Q (c.419G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.54 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
40
|
A
A
D
D
K
K
R
R
I
I
K
K
V
V
A
A
K
K
P
P
50
|
V
V
V
V
E
E
M
M
D
D
G
G
D
D
E
E
M
M
T
T
60
|
R
R
I
I
I
I
W
W
Q
Q
F
F
I
I
K
K
E
E
K
K
70
|
L
L
I
I
L
L
P
P
H
H
V
V
D
D
I
I
Q
Q
L
L
80
|
K
K
Y
Y
F
F
D
D
L
L
G
G
L
L
P
P
N
N
R
R
90
|
D
D
Q
Q
T
T
D
D
D
D
Q
Q
V
V
T
T
I
I
D
D
100
|
S
S
A
A
L
L
A
A
T
T
Q
Q
K
K
Y
Y
S
S
V
V
110
|
A
A
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
120
|
E
E
A
A
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
130
|
K
K
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
140
|
R
Q
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
150
|
E
E
P
P
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
160
|
L
L
V
V
P
P
G
G
W
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
K
R
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
A
A
T
T
D
D
F
F
V
V
A
A
D
D
R
R
A
A
190
|
G
G
T
T
F
F
K
K
M
M
V
V
F
F
T
T
P
P
K
K
200
|
D
D
G
G
S
S
G
G
V
V
K
K
E
E
W
W
E
E
V
V
210
|
Y
Y
N
N
F
F
P
P
A
A
G
G
G
G
V
V
G
G
M
M
220
|
G
G
M
M
Y
Y
N
N
T
T
D
D
E
E
S
S
I
I
S
S
230
|
G
G
F
F
A
A
H
H
S
S
C
C
F
F
Q
Q
Y
Y
A
A
240
|
I
I
Q
Q
K
K
K
K
W
W
P
P
L
L
Y
Y
M
M
S
S
250
|
T
T
K
K
N
N
T
T
I
I
L
L
K
K
A
A
Y
Y
D
D
260
|
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
I
I
270
|
F
F
D
D
K
K
H
H
Y
Y
K
K
T
T
D
D
F
F
D
D
280
|
K
K
N
N
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
L
L
290
|
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
V
V
L
L
K
K
300
|
S
S
S
S
G
G
G
G
F
F
V
V
W
W
A
A
C
C
K
K
310
|
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
I
I
320
|
L
L
A
A
Q
Q
G
G
F
F
G
G
S
S
L
L
G
G
L
L
330
|
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
G
G
340
|
K
K
T
T
I
I
E
E
A
A
E
E
A
A
A
A
H
H
G
G
350
|
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
E
E
H
H
Q
Q
360
|
K
K
G
G
R
R
P
P
T
T
S
S
T
T
N
N
P
P
I
I
370
|
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
L
L
380
|
E
E
H
H
R
R
G
G
K
K
L
L
D
D
G
G
N
N
Q
Q
390
|
D
D
L
L
I
I
R
R
F
F
A
A
Q
Q
M
M
L
L
E
E
400
|
K
K
V
V
C
C
V
V
E
E
T
T
V
V
E
E
S
S
G
G
410
|
A
A
M
M
T
T
K
K
D
D
L
L
A
A
G
G
C
C
I
I
420
|
H
H
G
G
L
L
S
S
N
N
V
V
K
K
L
L
N
N
E
E
430
|
H
H
F
F
L
L
N
N
T
T
T
T
D
D
F
F
L
L
D
D
440
|
T
T
I
I
K
K
S
S
N
N
L
L
D
D
R
R
A
A
L
L
450
|
G
G
R
R
Q
Q
S
L
L
E
E
H
H
H
H
H
H
H
H
H
460
|
H
H
H
H
H
H
H
-
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |||||||||
| TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | ||||||||||
| TF-1a cells | Bone marrow | Homo sapiens (Human) | CVCL_3608 | ||||||||||
| IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | NSG mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Somatic gain-of-function mutations in isocitrate dehydrogenases (IDH) 1 and 2 are found in multiple hematologic and solid tumors, leading to accumulation of the oncometabolite (R)-2-hydroxyglutarate (2HG). 2HG competitively inhibits alpha-ketoglutarate-dependent dioxygenases, including histone demethylases and methylcytosine dioxygenases of the TET family, causing epigenetic dysregulation and a block in cellular differentiation. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172K (c.515G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
40
|
-
A
-
D
-
K
-
R
-
I
-
K
-
V
-
A
-
K
-
P
50
|
-
V
-
V
-
E
-
M
-
D
-
G
-
D
-
E
-
M
-
T
60
|
-
R
-
I
-
I
-
W
-
Q
-
F
-
I
-
K
-
E
-
K
70
|
-
L
-
I
-
L
-
P
-
H
-
V
-
D
-
I
-
Q
-
L
80
|
-
K
-
Y
-
F
-
D
-
L
-
G
-
L
-
P
-
N
-
R
90
|
-
D
-
Q
-
T
-
D
-
D
-
Q
-
V
-
T
-
I
-
D
100
|
-
S
-
A
-
L
-
A
-
T
-
Q
-
K
-
Y
-
S
-
V
110
|
-
A
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
120
|
-
E
-
A
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
130
|
-
K
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
140
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
150
|
-
E
-
P
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
160
|
-
L
-
V
-
P
-
G
-
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
S
K
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
-
A
-
T
-
D
-
F
-
V
-
A
-
D
-
R
-
A
190
|
-
G
-
T
-
F
-
K
-
M
-
V
-
F
-
T
-
P
-
K
200
|
-
D
-
G
-
S
-
G
-
V
-
K
-
E
-
W
-
E
-
V
210
|
-
Y
-
N
-
F
-
P
-
A
-
G
-
G
-
V
-
G
-
M
220
|
-
G
-
M
-
Y
-
N
-
T
-
D
-
E
-
S
-
I
-
S
230
|
-
G
-
F
-
A
-
H
-
S
-
C
-
F
-
Q
-
Y
-
A
240
|
-
I
-
Q
-
K
-
K
-
W
-
P
-
L
-
Y
-
M
-
S
250
|
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
A
-
Y
-
D
260
|
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
-
I
270
|
-
F
-
D
-
K
-
H
-
Y
-
K
-
T
-
D
-
F
-
D
280
|
-
K
-
N
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
-
L
290
|
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
V
-
L
-
K
300
|
-
S
-
S
-
G
-
G
-
F
-
V
-
W
-
A
-
C
-
K
310
|
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
-
I
320
|
-
L
-
A
-
Q
-
G
-
F
-
G
-
S
-
L
-
G
-
L
330
|
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
-
G
340
|
-
K
-
T
-
I
-
E
-
A
-
E
-
A
-
A
-
H
-
G
350
|
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
E
-
H
-
Q
360
|
-
K
-
G
-
R
-
P
-
T
-
S
-
T
-
N
-
P
-
I
370
|
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
-
L
380
|
-
E
-
H
-
R
-
G
-
K
-
L
-
D
-
G
-
N
-
Q
390
|
-
D
-
L
-
I
-
R
-
F
-
A
-
Q
-
M
-
L
-
E
400
|
-
K
-
V
-
C
-
V
-
E
-
T
-
V
-
E
-
S
-
G
410
|
-
A
-
M
-
T
-
K
-
D
-
L
-
A
-
G
-
C
-
I
420
|
-
H
-
G
-
L
-
S
-
N
-
V
-
K
-
L
-
N
-
E
430
|
-
H
-
F
-
L
-
N
-
T
-
T
-
D
-
F
-
L
-
D
440
|
-
T
-
I
-
K
-
S
-
N
-
L
-
D
-
R
-
A
-
L
450
|
-
G
-
R
-
Q
-
S
-
L
-
E
-
H
-
H
-
H
-
H
460
|
-
H
-
H
-
H
-
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |||||||||
| TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | ||||||||||
| In Vivo Model | Acute myeloid leukemia xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
IC50 assay | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140Q (c.419G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.54 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
40
|
A
A
D
D
K
K
R
R
I
I
K
K
V
V
A
A
K
K
P
P
50
|
V
V
V
V
E
E
M
M
D
D
G
G
D
D
E
E
M
M
T
T
60
|
R
R
I
I
I
I
W
W
Q
Q
F
F
I
I
K
K
E
E
K
K
70
|
L
L
I
I
L
L
P
P
H
H
V
V
D
D
I
I
Q
Q
L
L
80
|
K
K
Y
Y
F
F
D
D
L
L
G
G
L
L
P
P
N
N
R
R
90
|
D
D
Q
Q
T
T
D
D
D
D
Q
Q
V
V
T
T
I
I
D
D
100
|
S
S
A
A
L
L
A
A
T
T
Q
Q
K
K
Y
Y
S
S
V
V
110
|
A
A
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
120
|
E
E
A
A
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
130
|
K
K
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
140
|
R
Q
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
150
|
E
E
P
P
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
160
|
L
L
V
V
P
P
G
G
W
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
K
R
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
A
A
T
T
D
D
F
F
V
V
A
A
D
D
R
R
A
A
190
|
G
G
T
T
F
F
K
K
M
M
V
V
F
F
T
T
P
P
K
K
200
|
D
D
G
G
S
S
G
G
V
V
K
K
E
E
W
W
E
E
V
V
210
|
Y
Y
N
N
F
F
P
P
A
A
G
G
G
G
V
V
G
G
M
M
220
|
G
G
M
M
Y
Y
N
N
T
T
D
D
E
E
S
S
I
I
S
S
230
|
G
G
F
F
A
A
H
H
S
S
C
C
F
F
Q
Q
Y
Y
A
A
240
|
I
I
Q
Q
K
K
K
K
W
W
P
P
L
L
Y
Y
M
M
S
S
250
|
T
T
K
K
N
N
T
T
I
I
L
L
K
K
A
A
Y
Y
D
D
260
|
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
I
I
270
|
F
F
D
D
K
K
H
H
Y
Y
K
K
T
T
D
D
F
F
D
D
280
|
K
K
N
N
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
L
L
290
|
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
V
V
L
L
K
K
300
|
S
S
S
S
G
G
G
G
F
F
V
V
W
W
A
A
C
C
K
K
310
|
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
I
I
320
|
L
L
A
A
Q
Q
G
G
F
F
G
G
S
S
L
L
G
G
L
L
330
|
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
G
G
340
|
K
K
T
T
I
I
E
E
A
A
E
E
A
A
A
A
H
H
G
G
350
|
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
E
E
H
H
Q
Q
360
|
K
K
G
G
R
R
P
P
T
T
S
S
T
T
N
N
P
P
I
I
370
|
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
L
L
380
|
E
E
H
H
R
R
G
G
K
K
L
L
D
D
G
G
N
N
Q
Q
390
|
D
D
L
L
I
I
R
R
F
F
A
A
Q
Q
M
M
L
L
E
E
400
|
K
K
V
V
C
C
V
V
E
E
T
T
V
V
E
E
S
S
G
G
410
|
A
A
M
M
T
T
K
K
D
D
L
L
A
A
G
G
C
C
I
I
420
|
H
H
G
G
L
L
S
S
N
N
V
V
K
K
L
L
N
N
E
E
430
|
H
H
F
F
L
L
N
N
T
T
T
T
D
D
F
F
L
L
D
D
440
|
T
T
I
I
K
K
S
S
N
N
L
L
D
D
R
R
A
A
L
L
450
|
G
G
R
R
Q
Q
S
L
L
E
E
H
H
H
H
H
H
H
H
H
460
|
H
H
H
H
H
H
H
-
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |||||||||
| TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | ||||||||||
| In Vivo Model | Acute myeloid leukemia xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
IC50 assay | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140G (c.418C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140W (c.418C>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140L (c.419G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172G (c.514A>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172W (c.514A>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172M (c.515G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [2] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172S (c.516G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Mechanism Description | Continuous daily enasidenib treatment was generally well tolerated and induced hematologic responses in patients for whom prior AML therapy had failed. Inducing differentiation of myeloblasts, not cytotoxicity, seems to drive the clinical efficacy of enasidenib. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [3] | ||||||||||||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172K (c.515G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
40
|
-
A
-
D
-
K
-
R
-
I
-
K
-
V
-
A
-
K
-
P
50
|
-
V
-
V
-
E
-
M
-
D
-
G
-
D
-
E
-
M
-
T
60
|
-
R
-
I
-
I
-
W
-
Q
-
F
-
I
-
K
-
E
-
K
70
|
-
L
-
I
-
L
-
P
-
H
-
V
-
D
-
I
-
Q
-
L
80
|
-
K
-
Y
-
F
-
D
-
L
-
G
-
L
-
P
-
N
-
R
90
|
-
D
-
Q
-
T
-
D
-
D
-
Q
-
V
-
T
-
I
-
D
100
|
-
S
-
A
-
L
-
A
-
T
-
Q
-
K
-
Y
-
S
-
V
110
|
-
A
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
120
|
-
E
-
A
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
130
|
-
K
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
140
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
150
|
-
E
-
P
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
160
|
-
L
-
V
-
P
-
G
-
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
S
K
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
-
A
-
T
-
D
-
F
-
V
-
A
-
D
-
R
-
A
190
|
-
G
-
T
-
F
-
K
-
M
-
V
-
F
-
T
-
P
-
K
200
|
-
D
-
G
-
S
-
G
-
V
-
K
-
E
-
W
-
E
-
V
210
|
-
Y
-
N
-
F
-
P
-
A
-
G
-
G
-
V
-
G
-
M
220
|
-
G
-
M
-
Y
-
N
-
T
-
D
-
E
-
S
-
I
-
S
230
|
-
G
-
F
-
A
-
H
-
S
-
C
-
F
-
Q
-
Y
-
A
240
|
-
I
-
Q
-
K
-
K
-
W
-
P
-
L
-
Y
-
M
-
S
250
|
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
A
-
Y
-
D
260
|
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
-
I
270
|
-
F
-
D
-
K
-
H
-
Y
-
K
-
T
-
D
-
F
-
D
280
|
-
K
-
N
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
-
L
290
|
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
V
-
L
-
K
300
|
-
S
-
S
-
G
-
G
-
F
-
V
-
W
-
A
-
C
-
K
310
|
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
-
I
320
|
-
L
-
A
-
Q
-
G
-
F
-
G
-
S
-
L
-
G
-
L
330
|
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
-
G
340
|
-
K
-
T
-
I
-
E
-
A
-
E
-
A
-
A
-
H
-
G
350
|
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
E
-
H
-
Q
360
|
-
K
-
G
-
R
-
P
-
T
-
S
-
T
-
N
-
P
-
I
370
|
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
-
L
380
|
-
E
-
H
-
R
-
G
-
K
-
L
-
D
-
G
-
N
-
Q
390
|
-
D
-
L
-
I
-
R
-
F
-
A
-
Q
-
M
-
L
-
E
400
|
-
K
-
V
-
C
-
V
-
E
-
T
-
V
-
E
-
S
-
G
410
|
-
A
-
M
-
T
-
K
-
D
-
L
-
A
-
G
-
C
-
I
420
|
-
H
-
G
-
L
-
S
-
N
-
V
-
K
-
L
-
N
-
E
430
|
-
H
-
F
-
L
-
N
-
T
-
T
-
D
-
F
-
L
-
D
440
|
-
T
-
I
-
K
-
S
-
N
-
L
-
D
-
R
-
A
-
L
450
|
-
G
-
R
-
Q
-
S
-
L
-
E
-
H
-
H
-
H
-
H
460
|
-
H
-
H
-
H
-
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [3] | ||||||||||||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R140K (c.418_419delCGinsAA) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Isocitrate dehydrogenase NADP 2 (IDH2) | [1] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R172K (c.515G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
40
|
-
A
-
D
-
K
-
R
-
I
-
K
-
V
-
A
-
K
-
P
50
|
-
V
-
V
-
E
-
M
-
D
-
G
-
D
-
E
-
M
-
T
60
|
-
R
-
I
-
I
-
W
-
Q
-
F
-
I
-
K
-
E
-
K
70
|
-
L
-
I
-
L
-
P
-
H
-
V
-
D
-
I
-
Q
-
L
80
|
-
K
-
Y
-
F
-
D
-
L
-
G
-
L
-
P
-
N
-
R
90
|
-
D
-
Q
-
T
-
D
-
D
-
Q
-
V
-
T
-
I
-
D
100
|
-
S
-
A
-
L
-
A
-
T
-
Q
-
K
-
Y
-
S
-
V
110
|
-
A
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
120
|
-
E
-
A
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
130
|
-
K
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
140
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
150
|
-
E
-
P
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
160
|
-
L
-
V
-
P
-
G
-
W
T
T
K
K
P
P
I
I
T
T
170
|
I
I
G
G
S
K
H
H
A
A
H
H
G
G
D
D
Q
Q
Y
Y
180
|
K
K
-
A
-
T
-
D
-
F
-
V
-
A
-
D
-
R
-
A
190
|
-
G
-
T
-
F
-
K
-
M
-
V
-
F
-
T
-
P
-
K
200
|
-
D
-
G
-
S
-
G
-
V
-
K
-
E
-
W
-
E
-
V
210
|
-
Y
-
N
-
F
-
P
-
A
-
G
-
G
-
V
-
G
-
M
220
|
-
G
-
M
-
Y
-
N
-
T
-
D
-
E
-
S
-
I
-
S
230
|
-
G
-
F
-
A
-
H
-
S
-
C
-
F
-
Q
-
Y
-
A
240
|
-
I
-
Q
-
K
-
K
-
W
-
P
-
L
-
Y
-
M
-
S
250
|
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
A
-
Y
-
D
260
|
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
-
I
270
|
-
F
-
D
-
K
-
H
-
Y
-
K
-
T
-
D
-
F
-
D
280
|
-
K
-
N
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
-
L
290
|
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
V
-
L
-
K
300
|
-
S
-
S
-
G
-
G
-
F
-
V
-
W
-
A
-
C
-
K
310
|
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
-
I
320
|
-
L
-
A
-
Q
-
G
-
F
-
G
-
S
-
L
-
G
-
L
330
|
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
-
G
340
|
-
K
-
T
-
I
-
E
-
A
-
E
-
A
-
A
-
H
-
G
350
|
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
E
-
H
-
Q
360
|
-
K
-
G
-
R
-
P
-
T
-
S
-
T
-
N
-
P
-
I
370
|
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
-
L
380
|
-
E
-
H
-
R
-
G
-
K
-
L
-
D
-
G
-
N
-
Q
390
|
-
D
-
L
-
I
-
R
-
F
-
A
-
Q
-
M
-
L
-
E
400
|
-
K
-
V
-
C
-
V
-
E
-
T
-
V
-
E
-
S
-
G
410
|
-
A
-
M
-
T
-
K
-
D
-
L
-
A
-
G
-
C
-
I
420
|
-
H
-
G
-
L
-
S
-
N
-
V
-
K
-
L
-
N
-
E
430
|
-
H
-
F
-
L
-
N
-
T
-
T
-
D
-
F
-
L
-
D
440
|
-
T
-
I
-
K
-
S
-
N
-
L
-
D
-
R
-
A
-
L
450
|
-
G
-
R
-
Q
-
S
-
L
-
E
-
H
-
H
-
H
-
H
460
|
-
H
-
H
-
H
-
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |||||||||
| TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | ||||||||||
| TF-1a cells | Bone marrow | Homo sapiens (Human) | CVCL_3608 | ||||||||||
| IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | NSG mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Somatic gain-of-function mutations in isocitrate dehydrogenases (IDH) 1 and 2 are found in multiple hematologic and solid tumors, leading to accumulation of the oncometabolite (R)-2-hydroxyglutarate (2HG). 2HG competitively inhibits alpha-ketoglutarate-dependent dioxygenases, including histone demethylases and methylcytosine dioxygenases of the TET family, causing epigenetic dysregulation and a block in cellular differentiation. | ||||||||||||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
