Drug Information
Drug (ID: DG01064) and It's Reported Resistant Information
| Name |
Prothionamide
|
||||
|---|---|---|---|---|---|
| Synonyms |
Protionamide; prothionamide; 14222-60-7; 2-propylpyridine-4-carbothioamide; Ektebin; Protionamid; Trevintix; Peteha; 2-Propyl-thioisonicotinamide; 2-Propylthioisonicotinamide; Tuberex; Protionamidum; Protionizina; Tebeform; 2-Propylisonicotinylthioamide; 4-Pyridinecarbothioamide, 2-propyl-; 2-Propyl-4-pyridinecarbothioamide; TH-1321; 2-Propyl-4-thiocarbamoylpyridine; RP 9778; Isonicotinamide, 2-propylthio-; 9778 R.P.; 1321 TH; UNII-76YOO33643; Prothionamidum; RP-9778; Protionamide (Prothionamide); MLS000042521; Protionamida; 76YOO33643; NSC-758962; NCGC00095164-01; SMR000047660; DSSTox_CID_25940; DSSTox_RID_81238; DSSTox_GSID_45940; Protionamidum [INN-Latin]; Protionamida [INN-Spanish]; Trevintix (TN); CAS-14222-60-7; Protionamide (INN); TH 1321; SR-05000001518; EINECS 238-093-7; BRN 0118164; Protion; Protionamide [INN:BAN:DCF]; Prothionamide (JP17); Opera_ID_999; Spectrum2_000019; Spectrum3_001964; SCHEMBL74572; BSPBio_003564; 5-22-02-00376 (Beilstein Handbook Reference); MLS001201789; MLS006011877; SPECTRUM1505316; SPBio_000057; CHEMBL1378024; DTXSID7045940; CHEBI:32066; KBio3_002911; Prothionamide, >=99% (HPLC); 2-Propyl-4-thiocarbamoyl pyridine; HMS1922D06; HMS2090J11; HMS2235M12; HMS3372K04; HMS3655O18; HMS3715G13; KUC109576N; Pharmakon1600-01505316; ALBB-010476; BCP13522; HY-B0306; KSC-27-052D; ZINC3874803; Tox21_111463; BBL010291; BDBM50499814; CCG-40049; MFCD00464119; NSC758962; s1881; STK366469; STL454225; AKOS005172678; Tox21_111463_1; 1321-TH; AC-4518; DB12667; KS-1282; MCULE-3817450515; NSC 758962; 2-propylpyridine-4-carbimidothioic acid; NCGC00095164-02; NCGC00095164-03; NCGC00095164-04; NCGC00095164-05; M860; SBI-0207058.P001; DB-042615; Protionamide 100 microg/mL in Acetonitrile; 4-Pyridinecarbothioamide, 2-propyl- (9CI); FT-0630412; P2302; SW199462-2; D01195; D88012; AB00393463-12; AB01093435-02; AB01093435_03; AB01093435_04; A807874; Q866657; Q-201638; SR-05000001518-1; SR-05000001518-2; SR-05000001518-3; BRD-K75360161-001-09-9
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
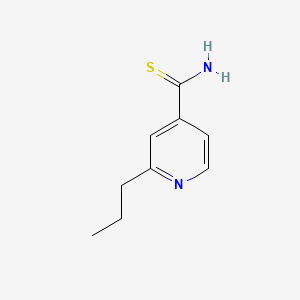
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[1]
|
||||
| Target | Bacterial Fatty acid synthetase I (Bact inhA) | INHA_MYCTU | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C9H12N2S
|
||||
| IsoSMILES |
CCCC1=NC=CC(=C1)C(=S)N
|
||||
| InChI |
1S/C9H12N2S/c1-2-3-8-6-7(9(10)12)4-5-11-8/h4-6H,2-3H2,1H3,(H2,10,12)
|
||||
| InChIKey |
VRDIULHPQTYCLN-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Enoyl-[acyl-carrier-protein] reductase [NADH] (INHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S94A |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.40 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
T
T
G
G
L
L
L
L
D
D
G
G
K
K
R
R
10
|
I
I
L
L
V
V
S
S
G
G
I
I
I
I
T
T
D
D
S
S
20
|
S
S
I
I
A
A
F
F
H
H
I
I
A
A
R
R
V
V
A
A
30
|
Q
Q
E
E
Q
Q
G
G
A
A
Q
Q
L
L
V
V
L
L
T
T
40
|
G
G
F
F
D
D
R
R
L
L
R
R
L
L
I
I
Q
Q
R
R
50
|
I
I
T
T
D
D
R
R
L
L
P
P
A
A
K
K
A
A
P
P
60
|
L
L
L
L
E
E
L
L
D
D
V
V
Q
Q
N
N
E
E
E
E
70
|
H
H
L
L
A
A
S
S
L
L
A
A
G
G
R
R
V
V
T
T
80
|
E
E
A
A
I
I
G
G
A
A
G
G
N
N
K
K
L
L
D
D
90
|
G
G
V
V
V
V
H
H
S
A
I
I
G
G
F
F
M
M
P
P
100
|
Q
Q
T
T
G
G
M
M
G
G
I
I
N
N
P
P
F
F
F
F
110
|
D
D
A
A
P
P
Y
Y
A
A
D
D
V
V
S
S
K
K
G
G
120
|
I
I
H
H
I
I
S
S
A
A
Y
Y
S
S
Y
Y
A
A
S
S
130
|
M
M
A
A
K
K
A
A
L
L
L
L
P
P
I
I
M
M
N
N
140
|
P
P
G
G
G
G
S
S
I
I
V
V
G
G
M
M
D
D
F
F
150
|
D
D
P
P
S
S
R
R
A
A
M
M
P
P
A
A
Y
Y
N
N
160
|
W
W
M
M
T
T
V
V
A
A
K
K
S
S
A
A
L
L
E
E
170
|
S
S
V
V
N
N
R
R
F
F
V
V
A
A
R
R
E
E
A
A
180
|
G
G
K
K
Y
Y
G
G
V
V
R
R
S
S
N
N
L
L
V
V
190
|
A
A
A
A
G
G
P
P
I
I
R
R
T
T
L
L
A
A
M
M
200
|
S
S
A
A
I
I
V
V
G
G
G
G
A
A
L
L
G
G
E
E
210
|
E
E
A
A
G
G
A
A
Q
Q
I
I
Q
Q
L
L
L
L
E
E
220
|
E
E
G
G
W
W
D
D
Q
Q
R
R
A
A
P
P
I
I
G
G
230
|
W
W
N
N
M
M
K
K
D
D
A
A
T
T
P
P
V
V
A
A
240
|
K
K
T
T
V
V
C
C
A
A
L
L
L
L
S
S
D
D
W
W
250
|
L
L
P
P
A
A
T
T
T
T
G
G
D
D
I
I
I
I
Y
Y
260
|
A
A
D
D
G
G
G
G
A
A
H
H
T
T
Q
Q
L
L
L
L
|
|||||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: Enoyl-[acyl-carrier-protein] reductase [NADH] (INHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G141E |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: Enoyl-[acyl-carrier-protein] reductase [NADH] (INHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.I194T |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.P28S |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L35R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G42D |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D56Y |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D58G |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.W69C |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.H102P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.C137R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y141N |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T186P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T189R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q246P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S266R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R279E |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S329P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.P334A |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.A341V |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.N345K |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.A352E |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.M372R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.C403Y |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.F480S |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.I161V |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G324R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q254P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S266R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S266R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.M373T |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L267V |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R239Q |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.S266R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q165P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q246R |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L446P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.V179F |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.A395D |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q254P |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G43S |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: FAD-containing monooxygenase EthA (ETHA) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.W69C |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: HTH-type transcriptional regulator EthR (ETHR) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.V152M |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
| Key Molecule: HTH-type transcriptional regulator EthR (ETHR) | [1] | ||||||||||||
| Resistant Disease | Mycolicibacterium smegmatis infection [ICD-11: 1B2Z.6] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R216C |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | Mycobacterium tuberculosis strain H37Rv ATCC27294 T | 83332 | |||||||||||
| Experiment for Molecule Alteration |
Sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing | ||||||||||||
| Mechanism Description | Notably, isoniazid is activated by the enzyme catalase-peroxidase, KatG, encoded by katG, whereas prothionamide is activated by the flavin monoxygenase, EthA, encoded by ethA. Mutations in katG and ethA are associated with individual isoniazid and prothionamide/ethionamide resistance, respectively. The ndh gene coding for NADH dehydrogenase, Ndh, was first identified as a new mechanism for INHR in Mycobacterium smegmatis. The mutations in ndh gene cause defects in the oxidation of NADH to NAD, which results in NADH accumulation and NAD depletion. The increased level of NADH inhibits the binding of isoniazid-NAD adduct to the active site of the InhA enzyme, which disturbs the regulation of enzyme activity and may cause co-resistance to isoniazid and prothionamide. EthR, a member of the TetR/CamR family, is a repressor of ethA. EthR regulates the transcription of ethA by coordinated octamerization on a 55-bp operator situated in the ethA-R intergenic region. Impeding EthR function leads to enhanced mycobacterial sensitivity to prothionamide, whereas mutations in ethR encoding a negative transcriptional regulator of the expression of EthA lead to prothionamide resistance. Finally, MshA, a member of the glycosyltransferase family, is a key enzyme involved in mycothiol biosynthesis in M. tuberculosis. Mutations in mshA coding MshA have been proposed to create a disturbance in prothionamide/ethionamide activation. | ||||||||||||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
