Drug Information
Drug (ID: DG00447) and It's Reported Resistant Information
| Name |
Cefpirome
|
||||
|---|---|---|---|---|---|
| Synonyms |
CEFPIROME; 84957-29-9; Cefpiroma; Cefpiromum; Cefrom; HR 810; UNII-S72Q2F09HY; cefpirome sulfate; CHEBI:3503; S72Q2F09HY; (6R,7R)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyiminoacetyl]amino]-3-(6,7-dihydro-5H-cyclopenta[b]pyridin-1-ium-1-ylmethyl)-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate; cefpirome sulphate; HR-810; (6R,7R)-7-{[(2Z)-2-(2-Amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-3-(6,7-dihydro-5H-cyclopenta[b]pyridinium-1-ylmethyl)-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate; 5H-Cyclopenta[b]pyridinium,1-[[(6R,7R)-7-[[(2Z)-(2-amino-4-thiazolyl)(methoxyimino)acetyl]amino]-2-carboxy-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-en-3-yl]methyl]-6,7-dihydro-, inner salt; Cefpiromum [Latin]; Cefpiroma [Spanish]; Cefpirome [INN:BAN]; NCGC00181339-01; Broact; Keiten; Cefir; Cefpirome (INN); Cefir (TN); HR-810 FREE BASE; SCHEMBL49406; MLS006010792; CHEMBL65794; DTXSID2048244; SCHEMBL22207951; C22H22N6O5S2; AKOS016013926; DB13682; (6R,7R)-7-(()-2-(2-Amino-4-thiazolyl)-2-methoxyiminoacetamido)-8-oxo-3-(2beta-trimethylenpyridinio)methyl)-5-thia-1-azabicyclo(4.2.0)oct-2-en-2-carboxylat; 1-(((6R,7R)-7-(2-(2-Amino-4-thiazolyl)glyoxylamino)-2-carboxy-8-oxo-5-thia-1-azabicyclo(4.2.0)oct-2-en-3-yl)methyl)-6,7-dihydro-5H-1-pyridinium hydroxide, inner salt; 5H-1-Pyrindinium, 1-((7-(((2-amino-4-thiazolyl)(methoxyimino)acetyl)amino)-2-carboxy-8-oxo-5-thia-1-azabicyclo(4.2.0)oct-2-en-3-yl)methyl)-6,7-dihydro-, hydroxide, inner salt, (6R-(6alpha,7beta(Z)))-; H828; SMR004701476; 98753-19-6; D07649; 1-{[(6R,7R)-2-carboxylato-7-{[(2Z)-1-hydroxy-2-(2-imino-2,3-dihydro-1,3-thiazol-4-yl)-2-(methoxyimino)ethylidene]amino}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-en-3-yl]methyl}-5H,6H,7H-cyclopenta[b]pyridin-1-ium; 5H-1-Pyrindinium, 1-((7-(((2-amino-4-thiazolyl)(methoxyimino)acetyl)amino)-2-carboxy-8-oxo-5-thia-1-azabicyclo(4.2.0)oct-2-en-3-yl)methyl)-5,6,7,8-tetrahydro-, hydroxide, inner salt, (6R-(6alpha,7beta(Z)))-; 7-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-3-(6,7-dihydro-5H-cyclopenta[b]pyridinium-1-ylmethyl)-3,4-didehydrocepham-4-carboxylic acid
Click to Show/Hide
|
||||
| Structure |
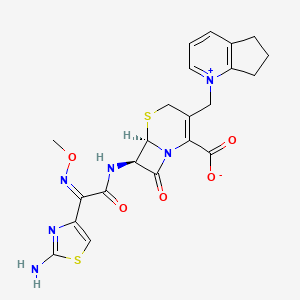
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(1 diseases)
|
||||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C22H22N6O5S2
|
||||
| IsoSMILES |
CO/N=C(/C1=CSC(=N1)N)\\C(=O)N[C@H]2[C@@H]3N(C2=O)C(=C(CS3)C[N+]4=CC=CC5=C4CCC5)C(=O)[O-]
|
||||
| InChI |
1S/C22H22N6O5S2/c1-33-26-15(13-10-35-22(23)24-13)18(29)25-16-19(30)28-17(21(31)32)12(9-34-20(16)28)8-27-7-3-5-11-4-2-6-14(11)27/h3,5,7,10,16,20H,2,4,6,8-9H2,1H3,(H3-,23,24,25,29,31,32)/b26-15-/t16-,20-/m1/s1
|
||||
| InChIKey |
DKOQGJHPHLTOJR-WHRDSVKCSA-N
|
||||
| PubChem CID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [1], [2], [3] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D240G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Citrobacter freundii 2526/96 | 546 | |||
| Escherichia coli isolates | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | We have reported recently the DNA sequence of another Beta-lactamase, CTX- M-15, from Indian enterobacterial isolates that were resistant to both cefotaxime and ceftazidime.CTX-M-15 has a single amino acid change [Asp-240-Gly (Ambler numbering)]7 compared with CTX-M-3. | |||
|
|
||||
| Key Molecule: Pyruvate decarboxylase 5 (PDC5) | [4], [3] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.R79Q+p.T105A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Pseudomonas aeruginosa isolates | 287 | |||
| Pseudomonas aeruginosa PAO1 | 208964 | |||
| Pseudomonas aeruginosa 12B | 287 | |||
| Pseudomonas aeruginosa kG2505 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; Etest method assay | |||
| Mechanism Description | Reduced susceptibility to imipenem, ceftazidime, and cefepime was observed only with recombinant P. aeruginosa strains expressing an AmpC Beta-lactamase that had an alanine residue at position 105.Recently, several ESACs have been described from Escherichia coli contributing to reduced susceptibility to imipenem. | |||
| Key Molecule: Pyruvate decarboxylase 3 (PDC3) | [4], [3] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.T97A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Pseudomonas aeruginosa isolates | 287 | |||
| Pseudomonas aeruginosa PAO1 | 208964 | |||
| Pseudomonas aeruginosa 12B | 287 | |||
| Pseudomonas aeruginosa kG2505 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; Etest method assay | |||
| Mechanism Description | Reduced susceptibility to imipenem, ceftazidime, and cefepime was observed only with recombinant P. aeruginosa strains expressing an AmpC Beta-lactamase that had an alanine residue at position 105.Recently, several ESACs have been described from Escherichia coli contributing to reduced susceptibility to imipenem. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
