Drug Information
Drug (ID: DG00403) and It's Reported Resistant Information
| Name |
Letrozole
|
||||
|---|---|---|---|---|---|
| Synonyms |
Femara; Femera; Letoval; Letrozol; Novartis Brand of Letrozole; CGS 20267; CGS-20267; FEM-345; Femara (TN); Letrozole [USAN:INN]; CGS 20267, Femara, Piroxicam, Letrozole; Letrozole (JAN/USP/INN); 1-[Bis-(4-cyanophenyl)methyl]-1,2,4-triazole; 1-[bis(4-cyanophenyl)methyl]-1,2,4-triazole; 4,4'-((1h-1,2,4-triazol-1-yl)methylene)dibenzonitrile; 4,4'-(1H-1,2,4-Triazol-1-ylmethylene)dibenzonitrile; 4,4'-(1H-1,2,4-triazol-1-yl-methylene)-bis(benzonitrile); 4,4'-(1H-1,2,4-triazol-1-ylmethanediyl)dibenzonitrile; 4,4'-(1H-1,2,4-triazol-1-ylmethylene)bis-Benzonitrile Letrozole; 4,4'-(1h-1,2,4-triazol-1-ylmethylene) bis-benzonitrile; 4,4'-(1h-1,2,4-triazol-1-ylmethylene)bis-benzonitrile; 4,4'-(1h-1,2,4-triazol-1-ylmethylene)bisbenzonitrile; 4-[(4-cyanophenyl)-(1,2,4-triazol-1-yl)methyl]benzonitrile
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
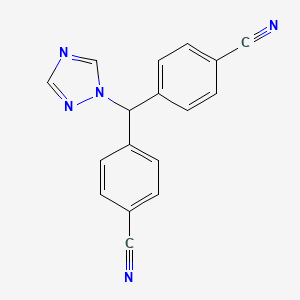
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(1 diseases)
[2]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[3]
|
||||
| Target | Aromatase (CYP19A1) | CP19A_HUMAN | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C17H11N5
|
||||
| IsoSMILES |
C1=CC(=CC=C1C#N)C(C2=CC=C(C=C2)C#N)N3C=NC=N3
|
||||
| InChI |
1S/C17H11N5/c18-9-13-1-5-15(6-2-13)17(22-12-20-11-21-22)16-7-3-14(10-19)4-8-16/h1-8,11-12,17H
|
||||
| InChIKey |
HPJKCIUCZWXJDR-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Deiodinase, iodothyronine type III, opposite strand (DIO3OS) | [3] | ||||||||||||
| Metabolic Type | Glucose metabolism | ||||||||||||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Four-week-old female athymic BALB/c nude mice, with tumor cells | Mice | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | Mechanistically, DIO3OS interacts with polypyrimidine tract binding protein 1 (PTBP1) and stabilizes the mRNA of lactate dehydrogenase A (LDHA) by protecting the integrity of its 3'UTR, and subsequently upregulates LDHA expression and activates glycolytic metabolism in AI-resistant breast cancer cells. Our findings highlight the role of lncRNA in regulating the key enzyme of glycolytic metabolism in response to endocrine therapies and the potential of targeting DIO3OS to reverse AI resistance in ER-positive breast cancer. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [2], [4] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y537S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.60 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.50 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
D
-
P
-
M
-
I
-
K
300
|
-
R
-
S
-
K
-
K
-
N
-
S
-
L
-
A
-
L
S
S
310
|
L
L
T
T
A
A
D
D
Q
Q
M
M
V
V
S
S
A
A
L
L
320
|
L
L
D
D
A
A
E
E
P
P
P
P
I
I
L
L
Y
Y
S
S
330
|
E
E
Y
Y
D
D
P
P
T
T
R
R
P
P
F
F
S
S
E
E
340
|
A
A
S
S
M
M
M
M
G
G
L
L
L
L
T
T
N
N
L
L
350
|
A
A
D
D
R
R
E
E
L
L
V
V
H
H
M
M
I
I
N
N
360
|
W
W
A
A
K
K
R
R
V
V
P
P
G
G
F
F
V
V
D
D
370
|
L
L
T
T
L
L
H
H
D
D
Q
Q
V
V
H
H
L
L
L
L
380
|
E
E
C
-
A
A
W
W
L
L
E
E
I
I
L
L
M
M
I
I
390
|
G
G
L
L
V
V
W
W
R
R
S
S
M
M
E
E
H
H
P
P
400
|
G
G
K
K
L
L
L
L
F
F
A
A
P
P
N
N
L
L
L
L
410
|
L
L
D
D
R
R
N
N
Q
Q
G
G
K
K
C
-
V
V
E
E
420
|
G
G
M
M
V
V
E
E
I
I
F
F
D
D
M
M
L
L
L
L
430
|
A
A
T
T
S
S
S
S
R
R
F
F
R
R
M
M
M
M
N
N
440
|
L
L
Q
Q
G
G
E
E
E
E
F
F
V
V
C
C
L
L
K
K
450
|
S
S
I
I
I
I
L
L
L
L
N
N
S
S
G
G
V
V
Y
Y
460
|
T
T
F
F
L
L
S
S
S
S
T
T
L
L
K
K
S
S
L
L
470
|
E
E
E
E
K
K
D
D
H
H
I
I
H
H
R
R
V
V
L
L
480
|
D
D
K
K
I
I
T
T
D
D
T
T
L
L
I
I
H
H
L
L
490
|
M
M
A
A
K
K
A
A
G
G
L
L
T
T
L
L
Q
Q
Q
Q
500
|
Q
Q
H
H
Q
Q
R
R
L
L
A
A
Q
Q
L
L
L
L
L
L
510
|
I
I
L
L
S
S
H
H
I
I
R
R
H
H
M
M
S
S
N
N
520
|
K
K
G
G
M
M
E
E
H
H
L
L
Y
Y
S
S
M
M
K
K
530
|
C
-
K
K
N
N
V
V
V
V
P
P
L
L
Y
S
D
D
L
L
540
|
L
L
L
L
E
E
M
M
L
L
D
D
A
A
H
H
R
R
L
L
550
|
H
H
A
A
P
P
T
T
S
S
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Droplet digital polymerase chain reaction assay | ||||||||||||
| Experiment for Drug Resistance |
Chest x-ray assay; Computed tomography assay; Magnetic resonance imaging assay; Positron emission tomography assay | ||||||||||||
| Mechanism Description | We have developed a ddPCR-based method for the sensitive detection and quantification of 4 representative ESR1 mutations, Y537S, Y537N, Y537C, and D538G, in 325 breast cancer specimens, in which 270 primary breast cancer and 55 MBC specimens. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [1] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D538G |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.60 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
V
-
D
-
L
-
G
-
T
-
E
290
|
-
N
-
L
-
Y
-
F
-
Q
-
S
-
N
-
A
-
M
-
K
300
|
-
R
-
S
-
K
-
K
-
N
-
S
-
L
-
A
-
L
S
S
310
|
L
L
T
T
A
A
D
D
Q
Q
M
M
V
V
S
S
A
A
L
L
320
|
L
L
D
D
A
A
E
E
P
P
P
P
I
I
L
L
Y
Y
S
S
330
|
E
E
Y
Y
D
D
P
P
T
T
R
R
P
P
F
F
S
S
E
E
340
|
A
A
S
S
M
M
M
M
G
G
L
L
L
L
T
T
N
N
L
L
350
|
A
A
D
D
R
R
E
E
L
L
V
V
H
H
M
M
I
I
N
N
360
|
W
W
A
A
K
K
R
R
V
V
P
P
G
G
F
F
V
V
D
D
370
|
L
L
T
T
L
L
H
H
D
D
Q
Q
V
V
H
H
L
L
L
L
380
|
E
E
C
C
A
A
W
W
L
L
E
E
I
I
L
L
M
M
I
I
390
|
G
G
L
L
V
V
W
W
R
R
S
S
M
M
E
E
H
H
P
P
400
|
G
G
K
K
L
L
L
L
F
F
A
A
P
P
N
N
L
L
L
L
410
|
L
L
D
D
R
R
N
N
Q
Q
G
G
K
K
C
C
V
V
E
E
420
|
G
G
M
M
V
V
E
E
I
I
F
F
D
D
M
M
L
L
L
L
430
|
A
A
T
T
S
S
S
S
R
R
F
F
R
R
M
M
M
M
N
N
440
|
L
L
Q
Q
G
G
E
E
E
E
F
F
V
V
C
C
L
L
K
K
450
|
S
S
I
I
I
I
L
L
L
L
N
N
S
S
G
G
V
V
Y
Y
460
|
T
T
F
F
L
L
S
S
S
S
T
T
L
L
K
K
S
S
L
L
470
|
E
E
E
E
K
K
D
D
H
H
I
I
H
H
R
R
V
V
L
L
480
|
D
D
K
K
I
I
T
T
D
D
T
T
L
L
I
I
H
H
L
L
490
|
M
M
A
A
K
K
A
A
G
G
L
L
T
T
L
L
Q
Q
Q
Q
500
|
Q
Q
H
H
Q
Q
R
R
L
L
A
A
Q
Q
L
L
L
L
L
L
510
|
I
I
L
L
S
S
H
H
I
I
R
R
H
H
M
M
S
S
N
N
520
|
K
K
G
G
M
M
E
E
H
H
L
L
Y
Y
S
S
M
M
K
K
530
|
C
C
K
K
N
N
V
V
V
V
P
P
L
L
Y
Y
D
G
L
L
540
|
L
L
L
L
E
E
M
M
L
L
D
D
A
A
H
H
R
R
L
L
550
|
H
H
A
A
P
P
T
T
S
S
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Sanger sequencing assay; dPCR assay | ||||||||||||
| Mechanism Description | Acquired mutations in the estrogen-receptor gene ESR1 can be a marker of resistance to aromatase inhibitors in metastatic breast cancer. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [2] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D538G |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.60 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
V
-
D
-
L
-
G
-
T
-
E
290
|
-
N
-
L
-
Y
-
F
-
Q
-
S
-
N
-
A
-
M
-
K
300
|
-
R
-
S
-
K
-
K
-
N
-
S
-
L
-
A
-
L
S
S
310
|
L
L
T
T
A
A
D
D
Q
Q
M
M
V
V
S
S
A
A
L
L
320
|
L
L
D
D
A
A
E
E
P
P
P
P
I
I
L
L
Y
Y
S
S
330
|
E
E
Y
Y
D
D
P
P
T
T
R
R
P
P
F
F
S
S
E
E
340
|
A
A
S
S
M
M
M
M
G
G
L
L
L
L
T
T
N
N
L
L
350
|
A
A
D
D
R
R
E
E
L
L
V
V
H
H
M
M
I
I
N
N
360
|
W
W
A
A
K
K
R
R
V
V
P
P
G
G
F
F
V
V
D
D
370
|
L
L
T
T
L
L
H
H
D
D
Q
Q
V
V
H
H
L
L
L
L
380
|
E
E
C
C
A
A
W
W
L
L
E
E
I
I
L
L
M
M
I
I
390
|
G
G
L
L
V
V
W
W
R
R
S
S
M
M
E
E
H
H
P
P
400
|
G
G
K
K
L
L
L
L
F
F
A
A
P
P
N
N
L
L
L
L
410
|
L
L
D
D
R
R
N
N
Q
Q
G
G
K
K
C
C
V
V
E
E
420
|
G
G
M
M
V
V
E
E
I
I
F
F
D
D
M
M
L
L
L
L
430
|
A
A
T
T
S
S
S
S
R
R
F
F
R
R
M
M
M
M
N
N
440
|
L
L
Q
Q
G
G
E
E
E
E
F
F
V
V
C
C
L
L
K
K
450
|
S
S
I
I
I
I
L
L
L
L
N
N
S
S
G
G
V
V
Y
Y
460
|
T
T
F
F
L
L
S
S
S
S
T
T
L
L
K
K
S
S
L
L
470
|
E
E
E
E
K
K
D
D
H
H
I
I
H
H
R
R
V
V
L
L
480
|
D
D
K
K
I
I
T
T
D
D
T
T
L
L
I
I
H
H
L
L
490
|
M
M
A
A
K
K
A
A
G
G
L
L
T
T
L
L
Q
Q
Q
Q
500
|
Q
Q
H
H
Q
Q
R
R
L
L
A
A
Q
Q
L
L
L
L
L
L
510
|
I
I
L
L
S
S
H
H
I
I
R
R
H
H
M
M
S
S
N
N
520
|
K
K
G
G
M
M
E
E
H
H
L
L
Y
Y
S
S
M
M
K
K
530
|
C
C
K
K
N
N
V
V
V
V
P
P
L
L
Y
Y
D
G
L
L
540
|
L
L
L
L
E
E
M
M
L
L
D
D
A
A
H
H
R
R
L
L
550
|
H
H
A
A
P
P
T
T
S
S
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Whole-genome sequencing assay | ||||||||||||
| Mechanism Description | Whole-exome and transcriptome analysis showed that six cases harbored mutations of ESR1 affecting its ligand-binding domain (LBD), all of whom had been treated with anti-estrogens and estrogen deprivation therapies. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [1] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y537N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Sanger sequencing assay; dPCR assay | ||||||||||||
| Mechanism Description | Acquired mutations in the estrogen-receptor gene ESR1 can be a marker of resistance to aromatase inhibitors in metastatic breast cancer. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [4] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y537N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Droplet digital polymerase chain reaction assay | ||||||||||||
| Experiment for Drug Resistance |
Chest x-ray assay; Computed tomography assay; Magnetic resonance imaging assay; Positron emission tomography assay | ||||||||||||
| Mechanism Description | We have developed a ddPCR-based method for the sensitive detection and quantification of 4 representative ESR1 mutations, Y537S, Y537N, Y537C, and D538G, in 325 breast cancer specimens, in which 270 primary breast cancer and 55 MBC specimens. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [4] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y537C |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Droplet digital polymerase chain reaction assay | ||||||||||||
| Experiment for Drug Resistance |
Chest x-ray assay; Computed tomography assay; Magnetic resonance imaging assay; Positron emission tomography assay | ||||||||||||
| Mechanism Description | We have developed a ddPCR-based method for the sensitive detection and quantification of 4 representative ESR1 mutations, Y537S, Y537N, Y537C, and D538G, in 325 breast cancer specimens, in which 270 primary breast cancer and 55 MBC specimens. | ||||||||||||
| Key Molecule: Estrogen receptor alpha (ESR1) | [2] | ||||||||||||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L536Q |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Whole-genome sequencing assay | ||||||||||||
| Mechanism Description | Whole-exome and transcriptome analysis showed that six cases harbored mutations of ESR1 affecting its ligand-binding domain (LBD), all of whom had been treated with anti-estrogens and estrogen deprivation therapies. | ||||||||||||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
