Drug Information
Drug (ID: DG00351) and It's Reported Resistant Information
| Name |
NSC141562
|
||||
|---|---|---|---|---|---|
| Synonyms |
NSC141562; NSC-141562; CHEMBL2003340; 1-[(2-methylquinolin-3-yl)methyl]-3,7-triaza-1-azoniatricyclo[3.3.1.13,7]decane, bromide
Click to Show/Hide
|
||||
| Structure |
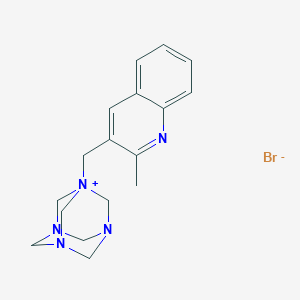
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[1]
|
||||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C17H22BrN5
|
||||
| IsoSMILES |
CC1=NC2=CC=CC=C2C=C1C[N+]34CN5CN(C3)CN(C5)C4.[Br-]
|
||||
| InChI |
1S/C17H22N5.BrH/c1-14-16(6-15-4-2-3-5-17(15)18-14)7-22-11-19-8-20(12-22)10-21(9-19)13-22;/h2-6H,7-13H2,1H3;1H/q+1;/p-1
|
||||
| InChIKey |
IYASYYOEAXFOPQ-UHFFFAOYSA-M
|
||||
| PubChem CID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MIR155 host gene (MIR155HG) | [1] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma multiforme | |||
| The Studied Tissue | Brain | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.58E-22 Fold-change: 2.18E+00 Z-score: 1.11E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | |
| Wnt/beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR; Microarray assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Wound healing assay; Transwell assay; MTT assay | |||
| Mechanism Description | miR155HG Is a Mesenchymal Transition-Associated Long Noncoding RNA, miR155-5p and miR155-3p Are key Derivatives of MIR155HG. miR155-5p or miR155-3p Targets Protocadherin 9 or 7, Respectively, Protocadherin 9 and 7 Function as Tumor Suppressor Genes by Inhibiting the Wnt/ beta-catenin signaling pathway. | |||
| Key Molecule: hsa-mir-155 | [1] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | |
| Wnt/beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Wound healing assay; Transwell assay; MTT assay | |||
| Mechanism Description | miR155HG Is a Mesenchymal Transition-Associated Long Noncoding RNA, miR155-5p and miR155-3p Are key Derivatives of MIR155HG. miR155-5p or miR155-3p Targets Protocadherin 9 or 7, Respectively, Protocadherin 9 and 7 Function as Tumor Suppressor Genes by Inhibiting the Wnt/ beta-catenin signaling pathway. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
