Drug Information
Drug (ID: DG00311) and It's Reported Resistant Information
| Name |
Cefprozil
|
||||
|---|---|---|---|---|---|
| Synonyms |
Arzimol; Brisoral; Cefprozilo; Cefprozilum; Cefzil; Cronocef; Procef; Serozil; Cefprozil anhydrous; BMY 28100; Cefprozil (INN); Cefprozil (TN); Cefprozilo [INN-Spanish]; Cefprozilum [INN-Latin]; Cefzil (TN); Procef (TN); (6R,7R)-7-[[(2R)-2-amino-2-(4-hydroxyphenyl)acetyl]amino]-8-oxo-3-[(E)-prop-1-enyl]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid; (6R,7R)-7-{[(2R)-2-amino-2-(4-hydroxyphenyl)acetyl]amino}-8-oxo-3-(prop-1-en-1-yl)-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
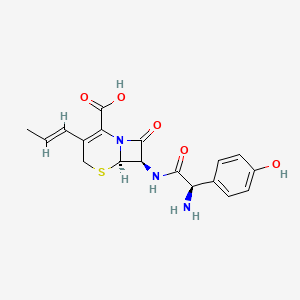
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(4 diseases)
|
||||
| Target | Bacterial Penicillin binding protein (Bact PBP) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C18H19N3O5S
|
||||
| IsoSMILES |
C/C=C/C1=C(N2[C@@H]([C@@H](C2=O)NC(=O)[C@@H](C3=CC=C(C=C3)O)N)SC1)C(=O)O
|
||||
| InChI |
1S/C18H19N3O5S/c1-2-3-10-8-27-17-13(16(24)21(17)14(10)18(25)26)20-15(23)12(19)9-4-6-11(22)7-5-9/h2-7,12-13,17,22H,8,19H2,1H3,(H,20,23)(H,25,26)/b3-2+/t12-,13-,17-/m1/s1
|
||||
| InChIKey |
WDLWHQDACQUCJR-ZAMMOSSLSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-10: Ear/mastoid process diseasess
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Penicillin-binding protein 2B (PBP2B) | [1], [2], [3] | |||
| Resistant Disease | Streptococcus pneumoniae infection [ICD-11: AA80.2] | |||
| Molecule Alteration | Missense mutation | p.T445A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae isolates | 1313 | ||
| Experiment for Molecule Alteration |
Genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | MICs for and PBP affinities of the strains correlated with the changes found in the PBP active binding sites.The PBP2b T445-A substitution found in all PISP and PRSP and one PSSP has been found in all low-level Beta-lactam-resistant pneumococci examined. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Penicillin-binding protein 2B (PBP2B) | [1], [2], [3] | |||
| Resistant Disease | Streptococcus pneumoniae infection [ICD-11: AA80.2] | |||
| Molecule Alteration | Missense mutation | p.T445A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae isolates | 1313 | ||
| Experiment for Molecule Alteration |
Genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | MICs for and PBP affinities of the strains correlated with the changes found in the PBP active binding sites.The PBP2b T445-A substitution found in all PISP and PRSP and one PSSP has been found in all low-level Beta-lactam-resistant pneumococci examined. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Penicillin-binding protein 2B (PBP2B) | [1], [2], [3] | |||
| Resistant Disease | Streptococcus pneumoniae infection [ICD-11: AA80.2] | |||
| Molecule Alteration | Missense mutation | p.T445A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae isolates | 1313 | ||
| Experiment for Molecule Alteration |
Genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | MICs for and PBP affinities of the strains correlated with the changes found in the PBP active binding sites.The PBP2b T445-A substitution found in all PISP and PRSP and one PSSP has been found in all low-level Beta-lactam-resistant pneumococci examined. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Penicillin-binding protein 2B (PBP2B) | [1], [2], [3] | |||
| Resistant Disease | Pneumocystis jirovecii infection [ICD-11: CA40.6] | |||
| Molecule Alteration | Missense mutation | p.T445A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae isolates | 1313 | ||
| Experiment for Molecule Alteration |
Genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | MICs for and PBP affinities of the strains correlated with the changes found in the PBP active binding sites.The PBP2b T445-A substitution found in all PISP and PRSP and one PSSP has been found in all low-level Beta-lactam-resistant pneumococci examined. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
