Drug Information
Drug (ID: DG00021) and It's Reported Resistant Information
| Name |
Cefalotin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Cefalothin; Cefalotina; Cefalotine; Cefalotinum; Cemastin; Cephalothinum; Cephalotin; Coaxin; Cefalotina fabra; Cephalothin Monosodium Salt; Averon-1; Cefalotin (BAN); Cefalotina [INN-Spanish]; Cefalotina fabra (TN); Cefalotine [INN-French]; Cefalotinum [INN-Latin]; Keflin (TN); (6R,7R)-3-(acetyloxymethyl)-8-oxo-7-[(2-thiophen-2-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid; (6R,7R)-3-[(acetyloxy)methyl]-8-oxo-7-[(2-thienylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid; 3-(Acetoxymethyl)-8-oxo-7-(2-(2-thienyl)acetamido)-5-thia-1-azabicyclo(4.2.0)oct-2-ene-2-carboxylic acid; 3-(Hydroxymethyl)-8-oxo-7-(2-(2-thienyl)acetamido)-5-thia-1-azabicyclo(4.2.0)oct-2-ene-2-carboxylic acid acetate; 3-ACETOXYMETHYL-8-OXO-7-(2-THIOPHEN-2-YL-ACETYLAMINO)-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-ENE-2-CARBOXYLIC ACID; 3-Acetoxymethyl-7-(2-thienylacetamido)-3-cephem-4-carboxylic acid; 5-Thia-1-azabicyclo(4.2.0)oct-2-ene-2-carboxylic acid, 3-(hydroxymethyl)-8-oxo-7-(2-(2-thienyl)acetamido)-, acetate (ester); 6R-trans-3-((Acetyloxy)methyl)-8-oxo-7-((2-thienylacetyl)amino)-5-thia-1-azabicyclo(4.2.0)-oct-2-ene-2-carboxylic acid; 7-(2-(2-Thienyl)acetylamido)cephalosporanic acid; 7-(2-Thienylacetamido)cephalosporanic acid; 7-(Thiophene-2-acetamido)cephalosporanic acid; 7-(Thiophene-2-acetamido)cephalosporin; 7beta-(thiophen-2-ylacetamido)-3-acetoxymethyl-3,4-didehydrocepham-4-carboxylic acid
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
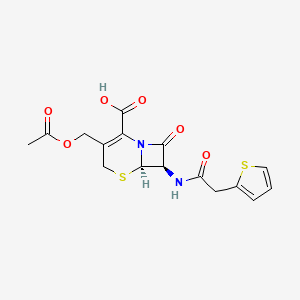
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(2 diseases)
[6]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[5]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[7]
|
||||
| Target | Bacterial DD-carboxypeptidase (Bact vanYB) | VANY_ENTFA | [2] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C16H16N2O6S2
|
||||
| IsoSMILES |
CC(=O)OCC1=C(N2[C@@H]([C@@H](C2=O)NC(=O)CC3=CC=CS3)SC1)C(=O)O
|
||||
| InChI |
1S/C16H16N2O6S2/c1-8(19)24-6-9-7-26-15-12(14(21)18(15)13(9)16(22)23)17-11(20)5-10-3-2-4-25-10/h2-4,12,15H,5-7H2,1H3,(H,17,20)(H,22,23)/t12-,15-/m1/s1
|
||||
| InChIKey |
XIURVHNZVLADCM-IUODEOHRSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Penicillin-binding protein 2C (PBP2C) | [5] | |||
| Resistant Disease | Lactobacillus casei infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | jk0412 cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Disk diffusion assay; Microdilution assay; E-Test assay | |||
| Mechanism Description | Six genes encoding putative high molecular weight penicillin-binding proteins (Pbp) are present in the genome of the beta-lactam-resistant strain?Corynebacterium jeikeium?K411. In this study, we show that?pbp2c, one of these six genes, is present in resistant strains of?Corynebacteriaceae?but absent from sensitive strains. The molecular study of the?pbp2c?locus from?C. jeikeium?and its heterologous expression in?Corynebacterium glutamicum?allowed us to show that Pbp2c confers high levels of beta-lactam resistance to the host and is under the control of a beta-lactam-induced regulatory system encoded by two adjacent genes,?jk0410?and?jk0411. The detection of this inducible resistance may require up to 48?h of incubation, particularly in?Corynebacterium amycolatum. Finally, the Pbp3c-expressing strains studied were resistant to all the beta-lactam antibiotics tested, including carbapenems, ceftaroline, and ceftobiprole. | |||
| Key Molecule: Penicillin-binding protein 2C (PBP2C) | [5] | |||
| Resistant Disease | Lactobacillus casei infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | cu1571 cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Disk diffusion assay; Microdilution assay; E-Test assay | |||
| Mechanism Description | Six genes encoding putative high molecular weight penicillin-binding proteins (Pbp) are present in the genome of the beta-lactam-resistant strain?Corynebacterium jeikeium?K411. In this study, we show that?pbp2c, one of these six genes, is present in resistant strains of?Corynebacteriaceae?but absent from sensitive strains. The molecular study of the?pbp2c?locus from?C. jeikeium?and its heterologous expression in?Corynebacterium glutamicum?allowed us to show that Pbp2c confers high levels of beta-lactam resistance to the host and is under the control of a beta-lactam-induced regulatory system encoded by two adjacent genes,?jk0410?and?jk0411. The detection of this inducible resistance may require up to 48?h of incubation, particularly in?Corynebacterium amycolatum. Finally, the Pbp6c-expressing strains studied were resistant to all the beta-lactam antibiotics tested, including carbapenems, ceftaroline, and ceftobiprole. | |||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [1], [2] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Escherichia coli JM101 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Beta-lactamases (Beta-lactamhydrolase, EC 3.5.2.6), responsible for most of the resistance to Beta-lactam antibiotics, are often plasmid mediated.The OXA-1 beta-lactamase gene is part of Tn2603, which is borne on the R plasmid RGN238. | |||
| Key Molecule: Beta-lactamase (BLA) | [8] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.Y104A+p.N110D+p.E175Q+p.S179A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Acinetobacter baumannii CIP70.10 | 470 | |||
| Klebsiella pneumoniae kP3 | 1290996 | |||
| Pseudomonas aeruginosa PU21 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | K. pneumoniae kP3 was resistant to all Beta-lactams, including carbapenems, and expressed the carbapenem-hydrolyzing Beta-lactamase OXA-181, which differs from OXA-48 by four amino acid substitutions. Compared to OXA-48, OXA-181 possessed a very similar hydrolytic profile. | |||
| Key Molecule: Beta-lactamase (BLA) | [9] | |||
| Resistant Disease | Pseudomonas aeruginosa infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa PAO1 | 208964 | ||
| Experiment for Molecule Alteration |
DNA sequencing and protein assay | |||
| Experiment for Drug Resistance |
Disk diffusion assay | |||
| Mechanism Description | P. aeruginosa harbors two naturally encoded Beta-lactamase genes, one of which encodes an inducible cephalosporinase and the other of which encodes a constitutively expressed oxacillinase. AmpC is a kind of cephalosporinase which lead to drug resistance. | |||
| Key Molecule: Beta-lactamase (BLA) | [2], [10] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D240G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli Gre-1 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | The first extended-spectrum Beta-lactamase (ESBL) of the CTX-M type (MEN-1/CTX-M-1) was reported at the beginning of the 1990s.CTX-M-27 differed from CTX-M-14 only by the substitution D240G and was the third CTX-M enzyme harbouring this mutation after CTX-M-15 and CTX-M-16. The Gly-240-harbouring enzyme CTX-M-27 conferred to Escherichia coli higher MICs of ceftazidime (MIC, 8 versus 1 mg/L) than did the Asp-240-harbouring CTX-M-14 enzyme. | |||
| Key Molecule: Beta-lactamase (BLA) | [2], [3], [4] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D240G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Citrobacter freundii 2526/96 | 546 | |||
| Escherichia coli isolates | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | We have reported recently the DNA sequence of another Beta-lactamase, CTX- M-15, from Indian enterobacterial isolates that were resistant to both cefotaxime and ceftazidime.CTX-M-15 has a single amino acid change [Asp-240-Gly (Ambler numbering)]7 compared with CTX-M-3. | |||
| Key Molecule: Beta-lactamase (BLA) | [7] | |||
| Resistant Disease | Rhodobacter sphaeroides infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Rhodopseudomonas sphaeroides strain DSM 160(Y) | 1063 | ||
| Rhodopseudomonas sphaeroides strain DSM158 | 1063 | |||
| Rhodopseudomonas sphaeroides strain DSM159 | 1063 | |||
| Experiment for Molecule Alteration |
Sodium dodecyl sulfate-PAGE assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Thirteen strains of the gram-negative, facultative phototrophic bacterium Rhodobacter sphaeroides were examined fro susceptibility to beta-lactam antibiotics. All strains were sensitive to the semisynthetic penicillins ampicillin, carbenicillin, oxacillin, cloxacillin, and methicillin, but 10 of the 13 strains were resistant to penicillin G, as well as a number of cephalosporins, such as cephalothin, cephapirin, and cephalosporin C. A beta-lactamase (EC 3.5.2.6) with strong cephalosporinase activity was detected in all of the resistant strains of R. sphaeroides. With strain Y-1 as a model, it was shown that the beta-lactamase was inducible by penicillin G, cephalosporin C, cephalothin, and to some minor extent, cephapirin. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Klebsiella pneumoniae ORI-1 strain harbored a ca. 140-kb nontransferable plasmid, pTk1, that conferred an extended-spectrum cephalosporin resistance profile antagonized by the addition of clavulanic acid, tazobactam, or imipenem. The gene for GES-1 (Guiana extended-spectrum beta-lactamase) was cloned, and its protein was expressed in Escherichia coli DH10B, where this pI-5. 8 beta-lactamase of a ca. 31-kDa molecular mass conferred resistance to oxyimino cephalosporins (mostly to ceftazidime). GES-1 is weakly related to the other plasmid-located Ambler class A extended-spectrum beta-lactamases (ESBLs). | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Beta-Lactam MICs for k. pneumoniae ORI-1 and Escherichia coli DH10B harboring either the natural plasmid pTk1 or the recombinant plasmid pC1 were somewhat similar and might indicate the presence of an ESBL. In all cases, the ceftazidime MICs were higher than those of cefotaxime and aztreonam. Beta-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Sensitive Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Antagonism |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Inhibition studies, as measured by IC50 values with benzylpenicillin as the substrate, showed that GES-1 was inhibited by clavulanic acid (5 uM) and tazobactam (2.5 uM) and strongly inhibited by imipenem (0.1 uM). Beta-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
