Molecule Information
General Information of the Molecule (ID: Mol01838)
| Name |
Granulocyte colony-stimulating factor receptor (CSF3R)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Granulocyte colony-stimulating factor receptor; G-CSF receptor; G-CSF-R; CD antigen CD114; CSF3R; GCSFR
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
CSF3R
|
||||
| Gene ID | |||||
| Location |
chr1:36,466,043-36,483,278[-]
|
||||
| Sequence |
MARLGNCSLTWAALIILLLPGSLEECGHISVSAPIVHLGDPITASCIIKQNCSHLDPEPQ
ILWRLGAELQPGGRQQRLSDGTQESIITLPHLNHTQAFLSCCLNWGNSLQILDQVELRAG YPPAIPHNLSCLMNLTTSSLICQWEPGPETHLPTSFTLKSFKSRGNCQTQGDSILDCVPK DGQSHCCIPRKHLLLYQNMGIWVQAENALGTSMSPQLCLDPMDVVKLEPPMLRTMDPSPE AAPPQAGCLQLCWEPWQPGLHINQKCELRHKPQRGEASWALVGPLPLEALQYELCGLLPA TAYTLQIRCIRWPLPGHWSDWSPSLELRTTERAPTVRLDTWWRQRQLDPRTVQLFWKPVP LEEDSGRIQGYVVSWRPSGQAGAILPLCNTTELSCTFHLPSEAQEVALVAYNSAGTSRPT PVVFSESRGPALTRLHAMARDPHSLWVGWEPPNPWPQGYVIEWGLGPPSASNSNKTWRME QNGRATGFLLKENIRPFQLYEIIVTPLYQDTMGPSQHVYAYSQEMAPSHAPELHLKHIGK TWAQLEWVPEPPELGKSPLTHYTIFWTNAQNQSFSAILNASSRGFVLHGLEPASLYHIHL MAASQAGATNSTVLTLMTLTPEGSELHIILGLFGLLLLLTCLCGTAWLCCSPNRKNPLWP SVPDPAHSSLGSWVPTIMEEDAFQLPGLGTPPITKLTVLEEDEKKPVPWESHNSSETCGL PTLVQTYVLQGDPRAVSTQPQSQSGTSDQVLYGQLLGSPTSPGPGHYLRCDSTQPLLAGL TPSPKSYENLWFQASPLGTLVTPAPSQEDDCVFGPLLNFPLLQGIRVHGMEALGSF Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Receptor for granulocyte colony-stimulating factor (CSF3), essential for granulocytic maturation. Plays a crucial role in the proliferation, differientation and survival of cells along the neutrophilic lineage. In addition it may function in some adhesion or recognition events at the cell surface.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.N610H (c.1828A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | C57/BL6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Sanger genomic DNA sequencing assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [2] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.T640N (c.1919C>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T17 cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| In Vivo Model | Balb/c bone marrow transplantation mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Sanger sequencing assay; Western blot analysis | |||
| Experiment for Drug Resistance |
Cytokine-independent growth assay | |||
| Disease Class: Atypical chronic myeloid leukemia [ICD-11: 2A41.1] | [3] | |||
| Sensitive Disease | Atypical chronic myeloid leukemia [ICD-11: 2A41.1] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.T615A (c.1843A>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
Direct sequencing assay | |||
| Mechanism Description | The missense mutation p.T615A (c.1843A>G) in gene CSF3R cause the sensitivity of Ruxolitinib by unusual activation of pro-survival pathway | |||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [4] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.T618I (c.1853C>T) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vivo Model | BALB/C nude mouse xenograft model | Mus musculus | ||
| Disease Class: Atypical chronic myeloid leukemia [ICD-11: 2A41.1] | [3] | |||
| Sensitive Disease | Atypical chronic myeloid leukemia [ICD-11: 2A41.1] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.T618I (c.1853C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
Direct sequencing assay | |||
| Mechanism Description | The missense mutation p.T618I (c.1853C>T) in gene CSF3R cause the sensitivity of Ruxolitinib by unusual activation of pro-survival pathway | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.N610S (c.1829A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | C57/BL6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Sanger genomic DNA sequencing assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.N610S (c.1829A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | C57/BL6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Sanger genomic DNA sequencing assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.N610H (c.1828A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | C57/BL6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Sanger genomic DNA sequencing assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
Preclinical Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ibrutinib/Ruxolitinib | |||
| Molecule Alteration | Nonsense | p.Q741* (c.2221C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | BTK signaling pathway | Inhibition | hsa04662 | |
| Experiment for Drug Resistance |
Trypan blue staining assay | |||
| Mechanism Description | G-CSFR mutants showed abnormal kinetics of canonical STAT3, STAT5 and MAPK phosphorylation, and aberrant activation of Bruton's Tyrosine Kinase (Btk). | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ibrutinib/Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.T618I (c.1853C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | BTK signaling pathway | Inhibition | hsa04662 | |
| Experiment for Drug Resistance |
Trypan blue staining assay | |||
| Mechanism Description | G-CSFR mutants showed abnormal kinetics of canonical STAT3, STAT5 and MAPK phosphorylation, and aberrant activation of Bruton's Tyrosine Kinase (Btk). | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.57E-03; Fold-change: -4.94E-01; Z-score: -1.30E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.69E-02; Fold-change: -2.12E-01; Z-score: -4.97E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
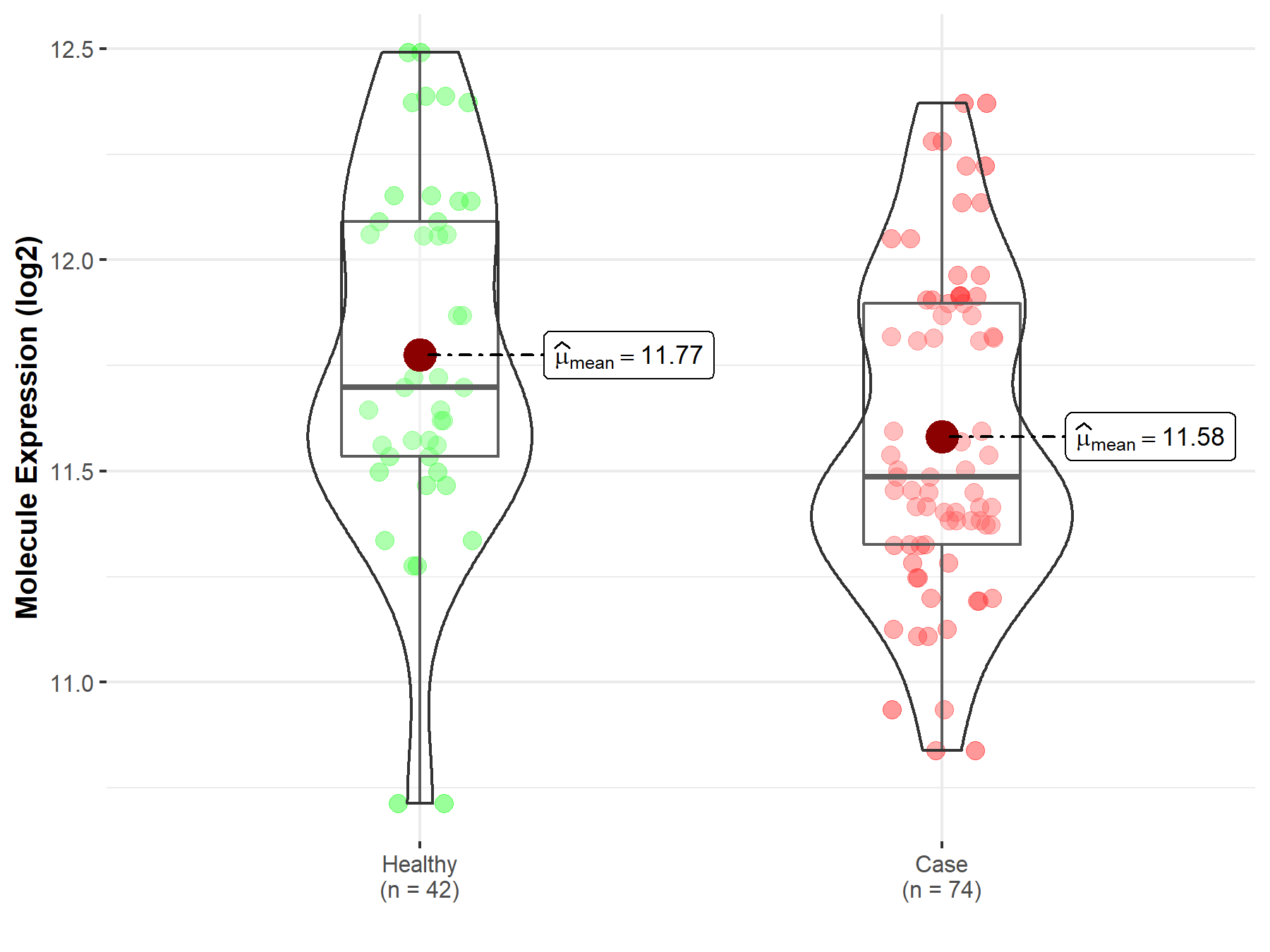
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

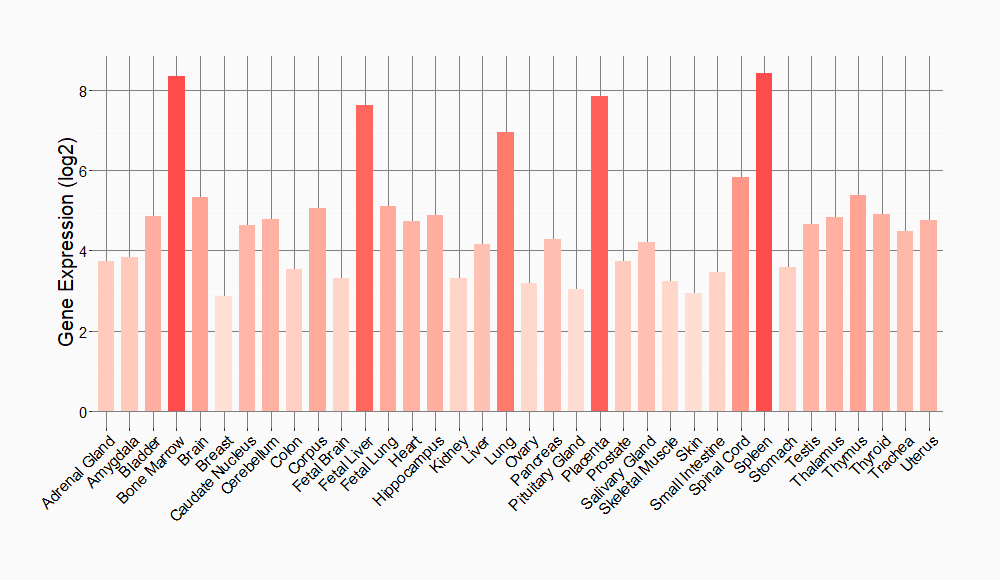
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
