Molecule Information
General Information of the Molecule (ID: Mol01826)
| Name |
PI3-kinase beta (PIK3CB)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
PI3-kinase subunit beta; PI3K-beta; PI3Kbeta; PtdIns-3-kinase subunit beta; Phosphatidylinositol 4;5-bisphosphate 3-kinase 110 kDa catalytic subunit beta; PtdIns-3-kinase subunit p110-beta; p110beta; PIK3CB; PIK3C1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PIK3CB
|
||||
| Gene ID | |||||
| Location |
chr3:138,652,698-138,834,928[-]
|
||||
| Sequence |
MCFSFIMPPAMADILDIWAVDSQIASDGSIPVDFLLPTGIYIQLEVPREATISYIKQMLW
KQVHNYPMFNLLMDIDSYMFACVNQTAVYEELEDETRRLCDVRPFLPVLKLVTRSCDPGE KLDSKIGVLIGKGLHEFDSLKDPEVNEFRRKMRKFSEEKILSLVGLSWMDWLKQTYPPEH EPSIPENLEDKLYGGKLIVAVHFENCQDVFSFQVSPNMNPIKVNELAIQKRLTIHGKEDE VSPYDYVLQVSGRVEYVFGDHPLIQFQYIRNCVMNRALPHFILVECCKIKKMYEQEMIAI EAAINRNSSNLPLPLPPKKTRIISHVWENNNPFQIVLVKGNKLNTEETVKVHVRAGLFHG TELLCKTIVSSEVSGKNDHIWNEPLEFDINICDLPRMARLCFAVYAVLDKVKTKKSTKTI NPSKYQTIRKAGKVHYPVAWVNTMVFDFKGQLRTGDIILHSWSSFPDELEEMLNPMGTVQ TNPYTENATALHVKFPENKKQPYYYPPFDKIIEKAAEIASSDSANVSSRGGKKFLPVLKE ILDRDPLSQLCENEMDLIWTLRQDCREIFPQSLPKLLLSIKWNKLEDVAQLQALLQIWPK LPPREALELLDFNYPDQYVREYAVGCLRQMSDEELSQYLLQLVQVLKYEPFLDCALSRFL LERALGNRRIGQFLFWHLRSEVHIPAVSVQFGVILEAYCRGSVGHMKVLSKQVEALNKLK TLNSLIKLNAVKLNRAKGKEAMHTCLKQSAYREALSDLQSPLNPCVILSELYVEKCKYMD SKMKPLWLVYNNKVFGEDSVGVIFKNGDDLRQDMLTLQMLRLMDLLWKEAGLDLRMLPYG CLATGDRSGLIEVVSTSETIADIQLNSSNVAAAAAFNKDALLNWLKEYNSGDDLDRAIEE FTLSCAGYCVASYVLGIGDRHSDNIMVKKTGQLFHIDFGHILGNFKSKFGIKRERVPFIL TYDFIHVIQQGKTGNTEKFGRFRQCCEDAYLILRRHGNLFITLFALMLTAGLPELTSVKD IQYLKDSLALGKSEEEALKQFKQKFDEALRESWTTKVNWMAHTVRKDYRS Click to Show/Hide
|
||||
| Function |
Phosphoinositide-3-kinase (PI3K) phosphorylates phosphatidylinositol derivatives at position 3 of the inositol ring to produce 3-phosphoinositides. Uses ATP and PtdIns(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Involved in the activation of AKT1 upon stimulation by G-protein coupled receptors (GPCRs) ligands such as CXCL12, sphingosine 1-phosphate, and lysophosphatidic acid. May also act downstream receptor tyrosine kinases. Required in different signaling pathways for stable platelet adhesion and aggregation. Plays a role in platelet activation signaling triggered by GPCRs, alpha-IIb/beta-3 integrins (ITGA2B/ ITGB3) and ITAM (immunoreceptor tyrosine-based activation motif)-bearing receptors such as GP6. Regulates the strength of adhesion of ITGA2B/ ITGB3 activated receptors necessary for the cellular transmission of contractile forces. Required for platelet aggregation induced by F2 (thrombin) and thromboxane A2 (TXA2). Has a role in cell survival. May have a role in cell migration. Involved in the early stage of autophagosome formation. Modulates the intracellular level of PtdIns3P (phosphatidylinositol 3-phosphate) and activates PIK3C3 kinase activity. May act as a scaffold, independently of its lipid kinase activity to positively regulate autophagy. May have a role in insulin signaling as scaffolding protein in which the lipid kinase activity is not required. May have a kinase-independent function in regulating cell proliferation and in clathrin-mediated endocytosis. Mediator of oncogenic signal in cell lines lacking PTEN. The lipid kinase activity is necessary for its role in oncogenic transformation. Required for the growth of ERBB2 and RAS driven tumors.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [1] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | PI3K pathway inhibitors | |||
| Molecule Alteration | Missense mutation | p.D1067Y (c.3199G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| SUM-52PE cells | Pleural effusion | Homo sapiens (Human) | CVCL_3425 | |
| Rat2 cells | Whole embryo | Rattus norvegicus (Rat) | CVCL_0513 | |
| HCC-1569 cells | Breast | Homo sapiens (Human) | CVCL_1255 | |
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | |
| A-498 cells | Kidney | Homo sapiens (Human) | CVCL_1056 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.82E-15; Fold-change: 2.44E-01; Z-score: 5.37E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.79E-11; Fold-change: 4.15E-01; Z-score: 1.11E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
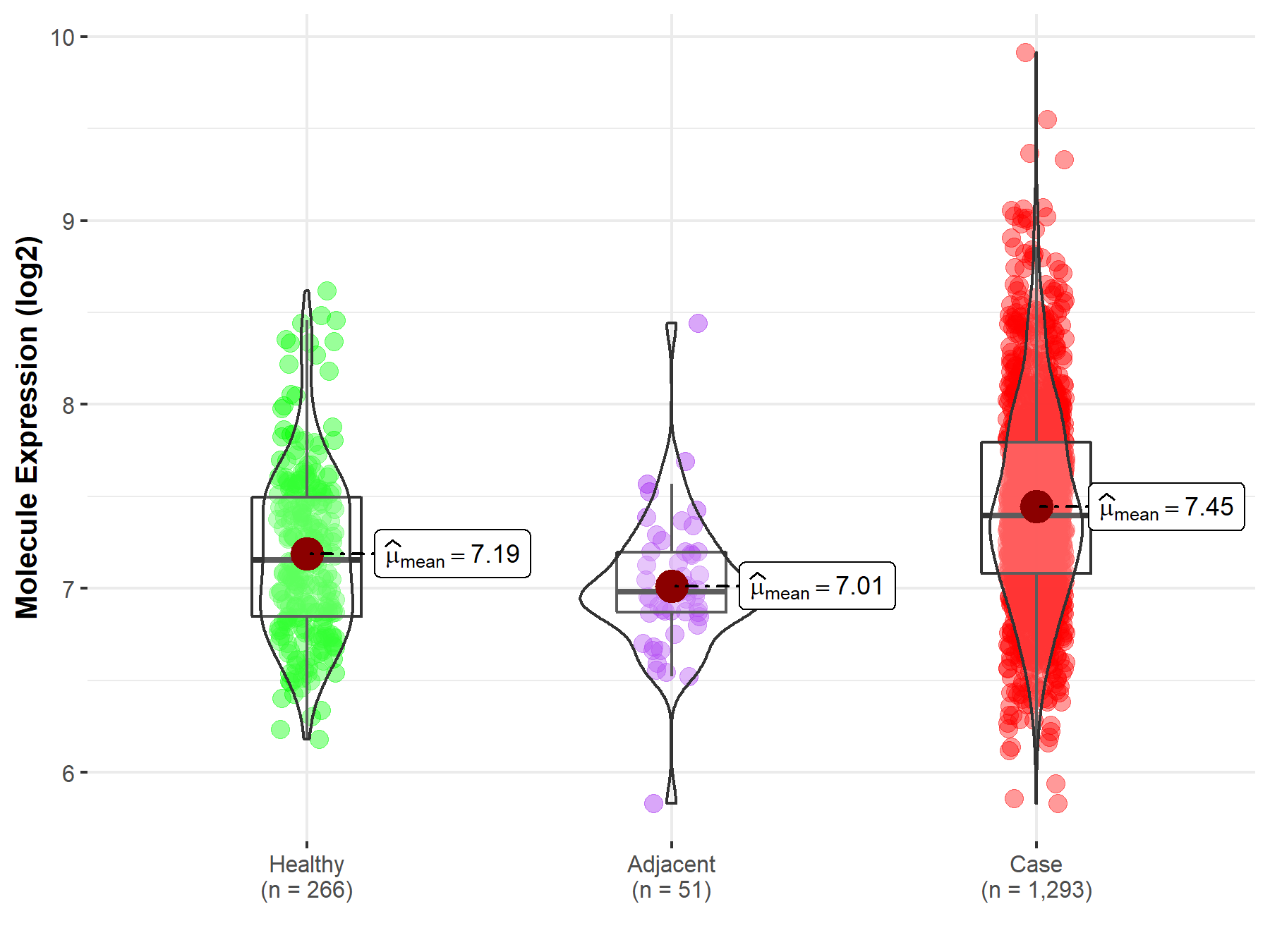
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

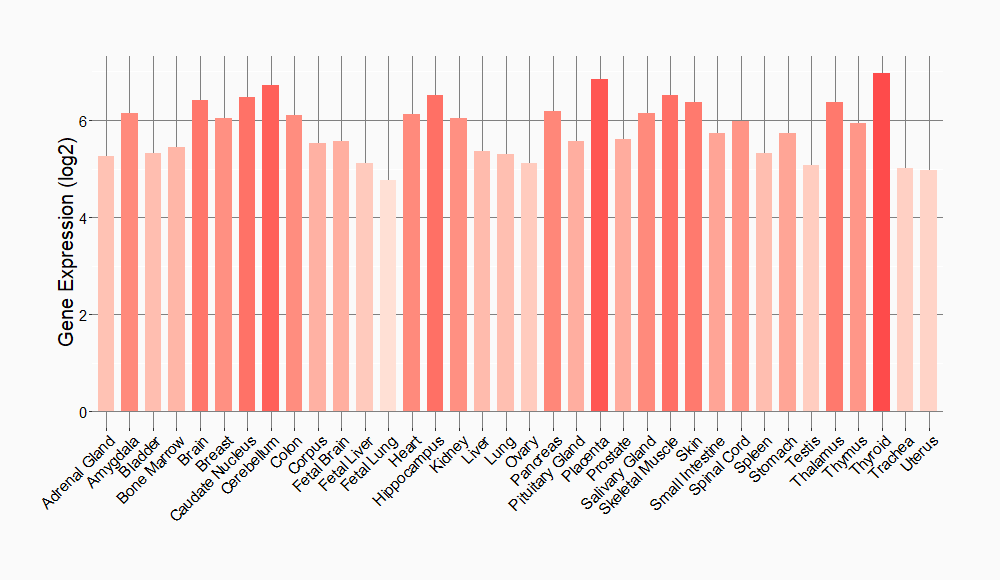
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
