Molecule Information
General Information of the Molecule (ID: Mol01407)
| Name |
hsa-mir-200b
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
microRNA 200b
Click to Show/Hide
|
||||
| Molecule Type |
Precursor miRNA
|
||||
| Gene Name |
MIR200B
|
||||
| Gene ID | |||||
| Location |
chr1:1167104-1167198[+]
|
||||
| Sequence |
CCAGCUCGGGCAGCCGUGGCCAUCUUACUGGGCAGCAUUGGAUGGAGUCAGGUCUCUAAU
ACUGCCUGGUAAUGAUGACGGCGGAGCCCUGCACG Click to Show/Hide
|
||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Precursor Accession | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
11 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [1] | |||
| Resistant Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Stomach adenocarcinoma | |||
| The Studied Tissue | Stomach | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.24E-02 Fold-change: -1.54E+00 Z-score: -3.43E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Fas/FasL signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The anti-apoptotic protein BCL2 and XIAP were upregulated, while the miR-200bc/429 cluster was downregulated in both SGC7901/VCR and A549/CDDP cells. miR-200bc/429 cluster might play an important role in the development of MDR in human gastric and lung cancer cell lines by targeting the anti-apoptotic genes BCL2 and XIAP. | |||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [3] | |||
| Resistant Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The transcription factor AP-2alpha functions as a tumor suppressor by regulating various genes that are involved in cell proliferation and apoptosis. Chemotherapeutic drugs including cisplatin induce post-transcriptionally endogenous AP-2alpha, which contributes to chemosensitivity by enhancing therapy-induced apoptosis. miR-200b/200c/429 family recognized the MRE in the 3' UTR of AP-2alpha gene and negatively regulated the expression of endogenous AP-2alpha proteins. | |||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [3] | |||
| Resistant Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEC-1A cells | Uterus | Homo sapiens (Human) | CVCL_0293 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The transcription factor AP-2alpha functions as a tumor suppressor by regulating various genes that are involved in cell proliferation and apoptosis. Chemotherapeutic drugs including cisplatin induce post-transcriptionally endogenous AP-2alpha, which contributes to chemosensitivity by enhancing therapy-induced apoptosis. miR-200b/200c/429 family recognized the MRE in the 3' UTR of AP-2alpha gene and negatively regulated the expression of endogenous AP-2alpha proteins. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Fas/FasL signaling pathway | Regulation | N.A. | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/CDDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The anti-apoptotic protein BCL2 and XIAP were upregulated, while the miR-200bc/429 cluster was downregulated in both SGC7901/VCR and A549/CDDP cells. miR-200bc/429 cluster might play an important role in the development of MDR in human gastric and lung cancer cell lines by targeting the anti-apoptotic genes BCL2 and XIAP. | |||
|
|
||||
| Disease Class: Tongue cancer [ICD-11: 2B62.0] | [4] | |||
| Resistant Disease | Tongue cancer [ICD-11: 2B62.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | SHH/GLI1 signaling pathway | Activation | hsa05217 | |
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 |
| SCC25 cells | Oral | Homo sapiens (Human) | CVCL_1682 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of BMI1 alters cell proliferation, apoptosis and stem cell self-renewal and correlates with the invasion and metastasis of several human cancers. BMI1 overexpression due to reduction of miR-200b and miR-15b may result in chemotherapy-induced EMT in TSCCs via these pathways. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [5] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| A2780CP cells | Ovary | Homo sapiens (Human) | CVCL_0135 | |
| HIOSE-80 cells | Ovary | Homo sapiens (Human) | CVCL_E274 | |
| OV119 cells | Ovary | Homo sapiens (Human) | N.A. | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-200b- and miR-200c-mediated downregulation of DNMTs may improve chemotherapeutic efficacy by increasing the sensitivity of cancer cells. | |||
|
|
||||
| Disease Class: Lung small cell carcinoma [ICD-11: 2C25.2] | [6] | |||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-200b was down-regulated in the resistant cells and enforced expression of miR-200b by miRNA mimics increased cell sensitivity. Overexpression of miR-200b led to the downregulation of ZEB2 at protein level. Luciferase reporter gene assay showed that 3'UTR ZEB2 activity was regulated by miR-200b. ZEB2 modulates drug resistance and is regulated by miR-200b. knockdown of ZEB2 increased cell sensitivity through increasing drug-induced cell apoptosis accompanied with S phase arrest. ZEB2 was regulated by miR-200b at protein level. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [2] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | Cetuximab | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | EGFR signaling pathway | Regulation | N.A. | |
| In Vitro Model | 253J BV cells | Bladder | Homo sapiens (Human) | CVCL_7937 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Pulse-labeling cells with [3H]thymidine | |||
| Mechanism Description | Members of the miR-200 family appear to control the EMT process and sensitivity to EGFR therapy, in bladder cancer cells and that expression of miR-200 is sufficient to restore EGFR dependency, at least in some of the mesenchymal bladder cancer cells. The targets of miR-200 include ERRFI-1, which is a novel regulator of EGFR-independent growth. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [7] | |||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | Six miRNAs (miR-192, 200b, 194, 424, 98 and 212) exhibited more than 2-fold changes in their expression levels, which were validated by qRT-PCR. The expression of three miRNAs (miR-200b, 194 and 212) was significantly down-regulated in SPC-A1/docetaxel cells, while the expression of other three miRNAs (miR-192, 424 and 98) was significantly up-regulated in SPC-A1/docetaxel cells (P < 0.01). | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [8] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| HDAC1/4/Sp1/miR200b/E2F3 signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | H1299/DTX cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| SPC-A1/DTX cells | Lung | Homo sapiens (Human) | CVCL_W217 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Histone deacetylase (HDAC) inhibitors could restore the expression of miR-200b and reverse chemoresistant phenotypes of docetaxel-resistant LAD cells. HDAC1/4 repression significantly increased miR-200b expression by upregulating histone-H3 acetylation level at the two miR-200b promoters partially via a Sp1-dependent pathway. Furthermore, silencing of HDAC1/4 suppressed cell proliferation, promoted cell apoptosis, induced G2/M cell cycle arrest and ultimately reversed in vitro and in vivo chemoresistance of docetaxel-resistant LAD cells, at least partially in a miR-200b-dependent manner. HDAC1/4 suppression-induced rescue of miR-200b contributed to downregulation of E2F3, survivin and Aurora-A, and upregulation of cleaved-caspase-3. HDAC1/4 levels in docetaxel-insensitive human LAD tissues, inversely correlated with miR-200b, were upregulated compared with docetaxel-sensitive tissues. Taken together, our findings suggest that the HDAC1/4/Sp1/miR-200b/E2F3 pathway is responsible for chemoresistance of docetaxel-resistant LAD cells. | |||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [9] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Bmi-1 is expressed at a high level in PCa. miR-200b plays a pivotal role in PCa at least in part via downregulation of the oncogene Bmi-1, inhibition of Bmi-1 enhanced the antitumor activity of docetaxel in PCa cells. | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [10] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | E2F3 was generally considered to increase cellular proliferation as a transcriptional activator through the G1/S transition, down-regulation of miR-200b could lead to E2F3 overexpression and in turn contribute to chemoresistance of lung adenocarcinoma cells to docetaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [12] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Erlotinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Hedgehog signaling pathway | Inhibition | hsa04340 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-200b and let-7c, inhibited the TGF-beta1-mediated resistance of NSCLC cells to erlotinib. | |||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [13] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Erlotinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| TGF-Beta/miR200/MIG6 signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | Calu3 cells | Lung | Homo sapiens (Human) | CVCL_0609 |
| H292 cells | Lung | Homo sapiens (Human) | CVCL_0455 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | |
| NCl-H226 cells | Lung | Homo sapiens (Human) | CVCL_1544 | |
| NCl-H1437 cells | Lung | Homo sapiens (Human) | CVCL_1472 | |
| H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| Calu6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| H1838 cells | Lung | Homo sapiens (Human) | CVCL_1499 | |
| H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR; RT-PCR | |||
| Experiment for Drug Resistance |
Alamar Blue assay | |||
| Mechanism Description | The Mig6-mediated reduction of EGFR occurs concomitantly with a TGFbeta-induced EMT-associated kinase switch of tumor cells to an AkT-activated state, thereby leading to an EGFR-independent phenotype that is refractory to EGFR TkI. the ratio of the expression levels of Mig6 and miR200c is highly correlated with EMT and resistance to erlotinib. Moreover, analyses of primary tumor xenografts of patient-derived lung and pancreatic cancers carrying wild type EGFR showed that the tumor Mig6(mRNA)/miR200 ratio is inversely correlated with response to erlotinib in vivo. | |||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [13] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | Erlotinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| TGF-Beta/miR200/MIG6 signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | Calu3 cells | Lung | Homo sapiens (Human) | CVCL_0609 |
| H292 cells | Lung | Homo sapiens (Human) | CVCL_0455 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | |
| NCl-H226 cells | Lung | Homo sapiens (Human) | CVCL_1544 | |
| NCl-H1437 cells | Lung | Homo sapiens (Human) | CVCL_1472 | |
| H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| Calu6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| H1838 cells | Lung | Homo sapiens (Human) | CVCL_1499 | |
| H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR; RT-PCR | |||
| Experiment for Drug Resistance |
Alamar Blue assay | |||
| Mechanism Description | The Mig6-mediated reduction of EGFR occurs concomitantly with a TGFbeta-induced EMT-associated kinase switch of tumor cells to an AkT-activated state, thereby leading to an EGFR-independent phenotype that is refractory to EGFR TkI. the ratio of the expression levels of Mig6 and miR200c is highly correlated with EMT and resistance to erlotinib. Moreover, analyses of primary tumor xenografts of patient-derived lung and pancreatic cancers carrying wild type EGFR showed that the tumor Mig6(mRNA)/miR200 ratio is inversely correlated with response to erlotinib in vivo. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cholangiocarcinoma [ICD-11: 2C12.0] | [14] | |||
| Sensitive Disease | Cholangiocarcinoma [ICD-11: 2C12.0] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | QBC939 cells | Bile duct | Homo sapiens (Human) | CVCL_6942 |
| TFk-1 cells | Bile duct | Homo sapiens (Human) | CVCL_2214 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
WST cell counting kit-8 | |||
| Mechanism Description | miR-200b/c influenced the tumourigenesis of cholangiocarcinoma cells including their tumour-initiating capacity, sphere formation, and drug resistance (like fluorouracil). We further found that miR-200b/c regulated migration and invasion capacities by directly targeting rho-kinase 2 and regulated tumorigenic properties by directly targeting SUZ12 (a subunit of a polycomb repressor complex). | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic carcinoma [ICD-11: 2C10.2] | [15] | |||
| Sensitive Disease | Pancreatic carcinoma [ICD-11: 2C10.2] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell metastasis | Inhibition | hsa05205 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| Chemosensitivity | Activation | hsa05207 | ||
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Colorimetric methylene blue assay; Flow cytometry assay | |||
| Mechanism Description | Forced expression of miR-200b induces CDH1 expression and promotes gemcitabine sensitivity in Capan-2 and Panc-1 cells. | |||
| Disease Class: Cholangiocarcinoma [ICD-11: 2C12.0] | [16] | |||
| Sensitive Disease | Cholangiocarcinoma [ICD-11: 2C12.0] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| KMCH-1 cells | Gallbladder | Homo sapiens (Human) | CVCL_7970 | |
| Mz-ChA-1 cells | Gallbladder | Homo sapiens (Human) | CVCL_6932 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Northern blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter 96 aqueous one solution cell proliferation assay | |||
| Mechanism Description | PTPN12 can bind and dephosphorylate the product ofoncogenes such as c-Abl or Src and inactivate the Raspathway. Thus, deregulation of PTPN12 expressionmay contribute to tumor cell survival and oncogenesis. In cells transfected with anti-miR-200b, PTPN12 ex-pression was increased to 132.2%+/-7.2% of controlafter 48 hours and 147.3%+/-12.8% of control after 72hours. Moreover, inhibition of miR-200b significantlyreduced the tyrosine phosphorylation of a downstreamtarget Src, a key mediator of tumor cell proliferation anddifferentiation. | |||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [17] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 |
| PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 | |
| AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | Re-expression of miR-200 in gemcitabine-resistant cells showed partial reversal of EMT characteristics as documented by increased expression of E-cadherin and decreased expression of vimentin, ZEB1, and slug. These results suggest that miR-200 family regulates the expression of ZEB1, slug, E-cadherin, and vimentin and that the re-expression of miR-200 could be useful for the reversal of EMT phenotype to mesenchymal-epithelial transition (MET). re-expression of miR-200 by transfection studies or treatment of gemcitabine-resistant cells with either DIM or isoflavone resulted in the down-regulation of ZEB1, slug, and vimentin, which was consistent with morphological reversal of EMT phenotype leading to epithelial morphology. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [18] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Intedanib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | |
| EBC-1 cells | Lung | Homo sapiens (Human) | CVCL_2891 | |
| LC-1/sq cells | Lung | Homo sapiens (Human) | CVCL_3008 | |
| LC-2/ad cells | Lung | Homo sapiens (Human) | CVCL_1373 | |
| Lk-2 cells | Lung | Homo sapiens (Human) | CVCL_1377 | |
| NCI-HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |
| PC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_S978 | |
| PC-10 cells | Lung | Homo sapiens (Human) | CVCL_7088 | |
| QG56 cells | Lung | Homo sapiens (Human) | CVCL_6943 | |
| RERF-LCkJ cells | Lung | Homo sapiens (Human) | CVCL_1654 | |
| SQ5 cells | Lung | Homo sapiens (Human) | CVCL_8273 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-200b and miR-141 associated with epithelial-mesenchymal transition (EMT) are predictive biomarkers and therapeutic targets of nintedanib in NSCLC cells. nintedanib inhibited EMT and reversed the resistance to EGFR-TkI with TGF-beta-induced EMT through miR-200 family induction in NSCLC cells. low expression of miR-200b and miR-141, resulting in high level of ZEB1 and low level of E-cadherin, was associated with the resistance to nintedanib in NSCLC cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [19] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 |
| Malme3M cells | Skin | Homo sapiens (Human) | CVCL_1438 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-326, which forms a negative feedback regulatory loop with HDAC3, regulates the invasion and the metastatic potential of cancer cells and tumor-induced angiogenesis in response to anti-cancer drugs. miR-200b, miR-217, and miR-335, which form a positive feedback loop with HDAC3, confer sensitivity to anti-cancer drugs. We show that CAGE, reported to form a feedback loop with miR-200b, serves as a downstream target of HDAC3 and miR-326. In this study, we show that the regulation of the miR-326/HDAC3 axis can be employed for the development of anti-cancer therapeutics. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [1] | |||
| Resistant Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Resistant Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Fas/FasL signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The anti-apoptotic protein BCL2 and XIAP were upregulated, while the miR-200bc/429 cluster was downregulated in both SGC7901/VCR and A549/CDDP cells. miR-200bc/429 cluster might play an important role in the development of MDR in human gastric and lung cancer cell lines by targeting the anti-apoptotic genes BCL2 and XIAP. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Fas/FasL signaling pathway | Regulation | N.A. | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/CDDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The anti-apoptotic protein BCL2 and XIAP were upregulated, while the miR-200bc/429 cluster was downregulated in both SGC7901/VCR and A549/CDDP cells. miR-200bc/429 cluster might play an important role in the development of MDR in human gastric and lung cancer cell lines by targeting the anti-apoptotic genes BCL2 and XIAP. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [20] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Curcumin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HepJ5 cells | Liver | Homo sapiens (Human) | CVCL_RW48 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The overexpression ofmiR-200a/b in HepJ5 cells conferred enhanced resistance tocurcumin treatment compared with the control cells. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [19] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Celastrol | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 |
| Malme3M cells | Skin | Homo sapiens (Human) | CVCL_1438 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-326, which forms a negative feedback regulatory loop with HDAC3, regulates the invasion and the metastatic potential of cancer cells and tumor-induced angiogenesis in response to anti-cancer drugs. miR-200b, miR-217, and miR-335, which form a positive feedback loop with HDAC3, confer sensitivity to anti-cancer drugs. We show that CAGE, reported to form a feedback loop with miR-200b, serves as a downstream target of HDAC3 and miR-326. In this study, we show that the regulation of the miR-326/HDAC3 axis can be employed for the development of anti-cancer therapeutics. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

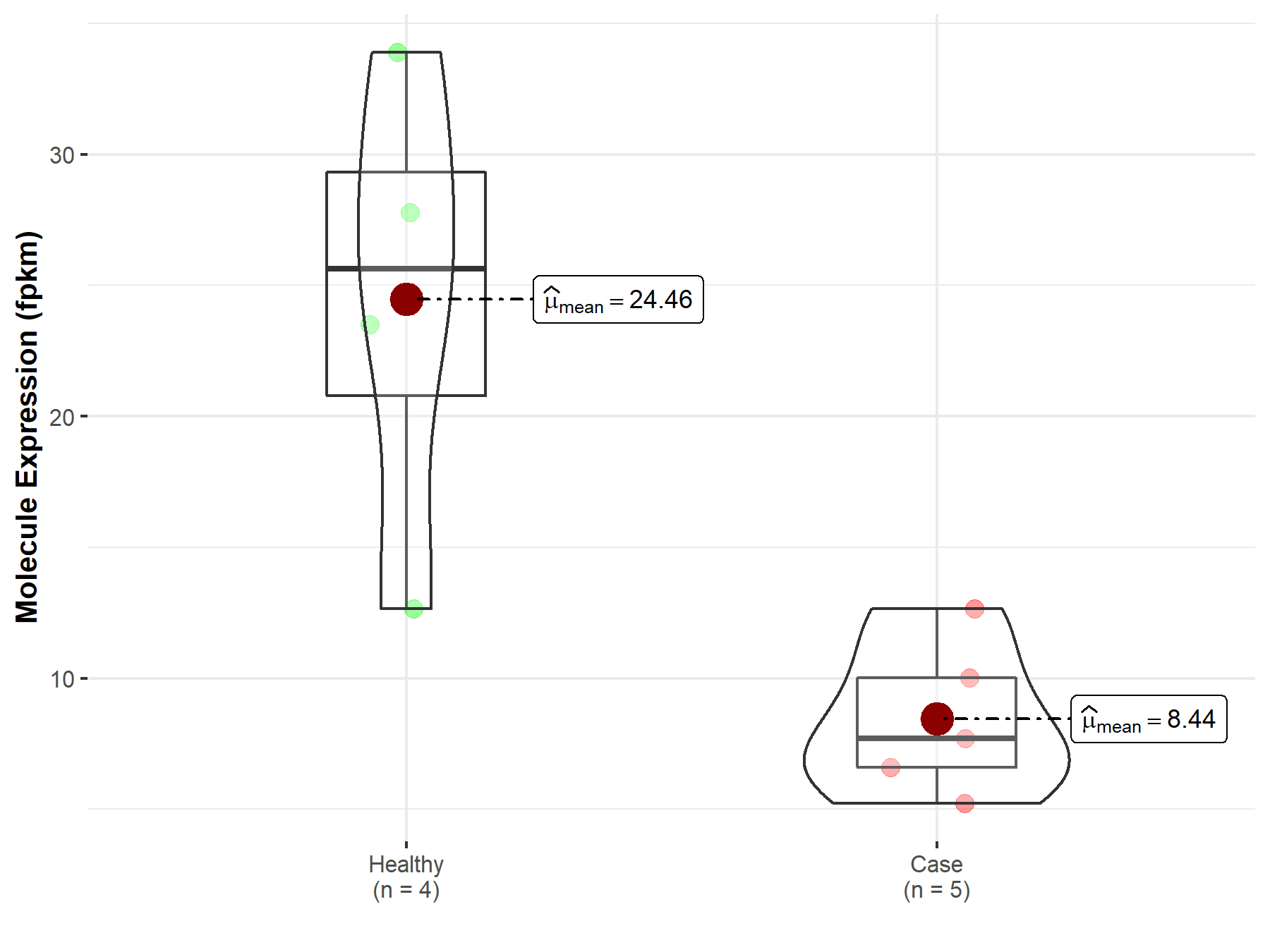
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Stomach | |
| The Specified Disease | Stomach adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.93E-04; Fold-change: 1.65E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
