Molecule Information
General Information of the Molecule (ID: Mol00768)
| Name |
Alpha-enolase (ENO1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
2-phospho-D-glycerate hydro-lyase; C-myc promoter-binding protein; Enolase 1; MBP-1; MPB-1; Non-neural enolase; NNE; Phosphopyruvate hydratase; Plasminogen-binding protein; ENO1L1; MBPB1; MPB1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ENO1
|
||||
| Gene ID | |||||
| Location |
chr1:8861000-8879190[-]
|
||||
| Sequence |
MSILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGK
GVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCK AGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFRE AMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKV VIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDD WGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLA QANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSK AKFAGRNFRNPLAK Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Glycolytic enzyme the catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate. In addition to glycolysis, involved in various processes such as growth control, hypoxia tolerance and allergic responses. May also function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons. Stimulates immunoglobulin production.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.20E-06 Fold-change: 6.37E-02 Z-score: 4.72E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Hk2, PFkB3, ENO-1, and PkM-2 are the main enzymes of glycolysis and their expression is upregulated in TAMR cells. Hk2 is also involved in the activation of pro-survival autophagy. ENO-1 plays an important role by inhibiting apoptosis via downregulation of c-Myc. In mitochondria, PDk4 phosphorylation regulate Pyruvate dehydrogenase of PDC. NSD2 activates Hk-2, G6PD, and TIGAR expression and upregulates the PPP pathway. PPP produces NADH and Ribulose 5-Phosphate, a substrate for nucleotide biosynthesis. NADH reduces ROS and inhibits apoptosis. LDHA overexpression helps in aerobic glycolysis and indirectly promotes autophagy. To reduce the concentration of lactate in the cells, MCT expression is increased, which facilitates the efflux of lactate and cell survival. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.20E-06; Fold-change: 3.92E-01; Z-score: 3.66E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.29E-01; Fold-change: -5.41E-02; Z-score: -4.99E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
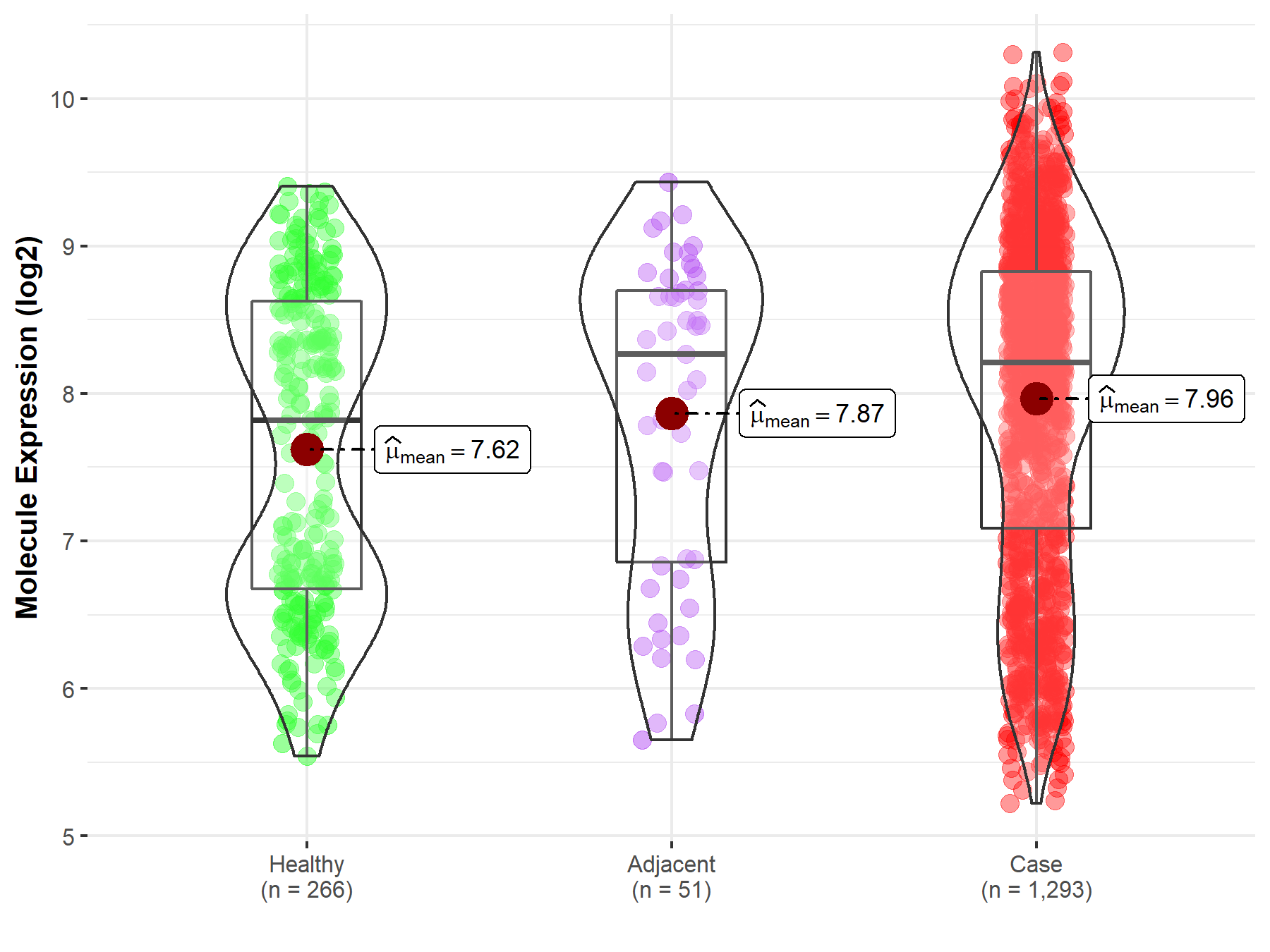
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

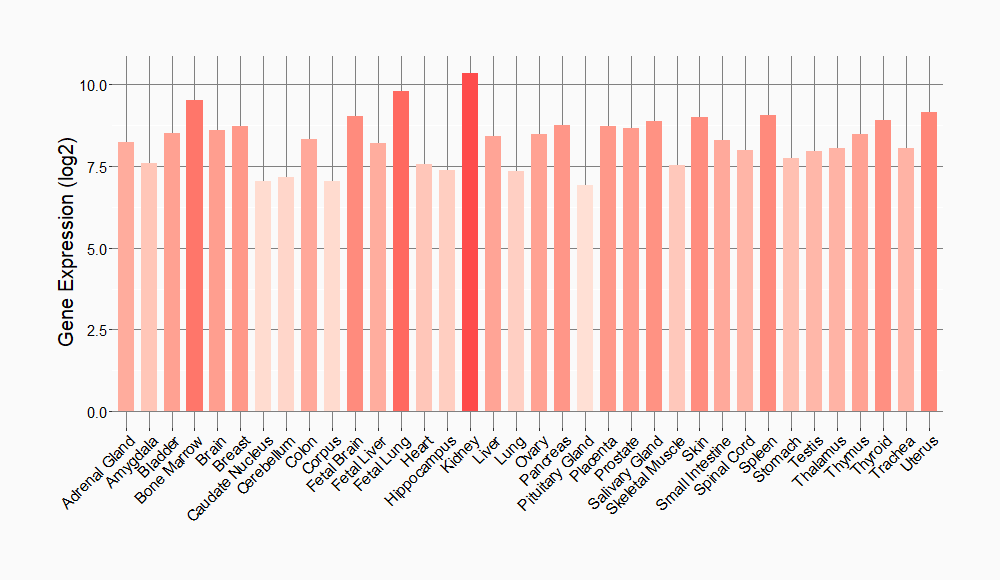
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
