Molecule Information
General Information of the Molecule (ID: Mol00714)
| Name |
Tyrosine-protein kinase Yes (YES1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Proto-oncogene c-Yes; p61-Yes; YES
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
YES1
|
||||
| Gene ID | |||||
| Location |
chr18:721588-812546[-]
|
||||
| Sequence |
MGCIKSKENKSPAIKYRPENTPEPVSTSVSHYGAEPTTVSPCPSSSAKGTAVNFSSLSMT
PFGGSSGVTPFGGASSSFSVVPSSYPAGLTGGVTIFVALYDYEARTTEDLSFKKGERFQI INNTEGDWWEARSIATGKNGYIPSNYVAPADSIQAEEWYFGKMGRKDAERLLLNPGNQRG IFLVRESETTKGAYSLSIRDWDEIRGDNVKHYKIRKLDNGGYYITTRAQFDTLQKLVKHY TEHADGLCHKLTTVCPTVKPQTQGLAKDAWEIPRESLRLEVKLGQGCFGEVWMGTWNGTT KVAIKTLKPGTMMPEAFLQEAQIMKKLRHDKLVPLYAVVSEEPIYIVTEFMSKGSLLDFL KEGDGKYLKLPQLVDMAAQIADGMAYIERMNYIHRDLRAANILVGENLVCKIADFGLARL IEDNEYTARQGAKFPIKWTAPEAALYGRFTIKSDVWSFGILQTELVTKGRVPYPGMVNRE VLEQVERGYRMPCPQGCPESLHELMNLCWKKDPDERPTFEYIQSFLEDYFTATEPQYQPG ENL Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Non-receptor protein tyrosine kinase that is involved in the regulation of cell growth and survival, apoptosis, cell-cell adhesion, cytoskeleton remodeling, and differentiation. Stimulation by receptor tyrosine kinases (RTKs) including EGRF, PDGFR, CSF1R and FGFR leads to recruitment of YES1 to the phosphorylated receptor, and activation and phosphorylation of downstream substrates. Upon EGFR activation, promotes the phosphorylation of PARD3 to favor epithelial tight junction assembly. Participates in the phosphorylation of specific junctional components such as CTNND1 by stimulating the FYN and FER tyrosine kinases at cell-cell contacts. Upon T-cell stimulation by CXCL12, phosphorylates collapsin response mediator protein 2/DPYSL2 and induces T-cell migration. Participates in CD95L/FASLG signaling pathway and mediates AKT-mediated cell migration. Plays a role in cell cycle progression by phosphorylating the cyclin-dependent kinase 4/CDK4 thus regulating the G1 phase. Also involved in G2/M progression and cytokinesis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [1] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Prostate cancer [ICD-11: 2C82] | |||
| The Specified Disease | Prostate cancer | |||
| The Studied Tissue | Prostate | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.64E-01 Fold-change: 3.34E-03 Z-score: 1.72E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | PC3/TXR cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC and PI Flow cytometry assay | |||
| Mechanism Description | Overexpression of miR199a inhibited PTX resistance. YES1 was a target of miR199a, and overexpression of YES1 reversed the effect of miR199a in suppressing PTX resistance. In vivo, miR199a increased tumor PTX sensitivity. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.64E-01; Fold-change: 2.55E-01; Z-score: 4.70E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
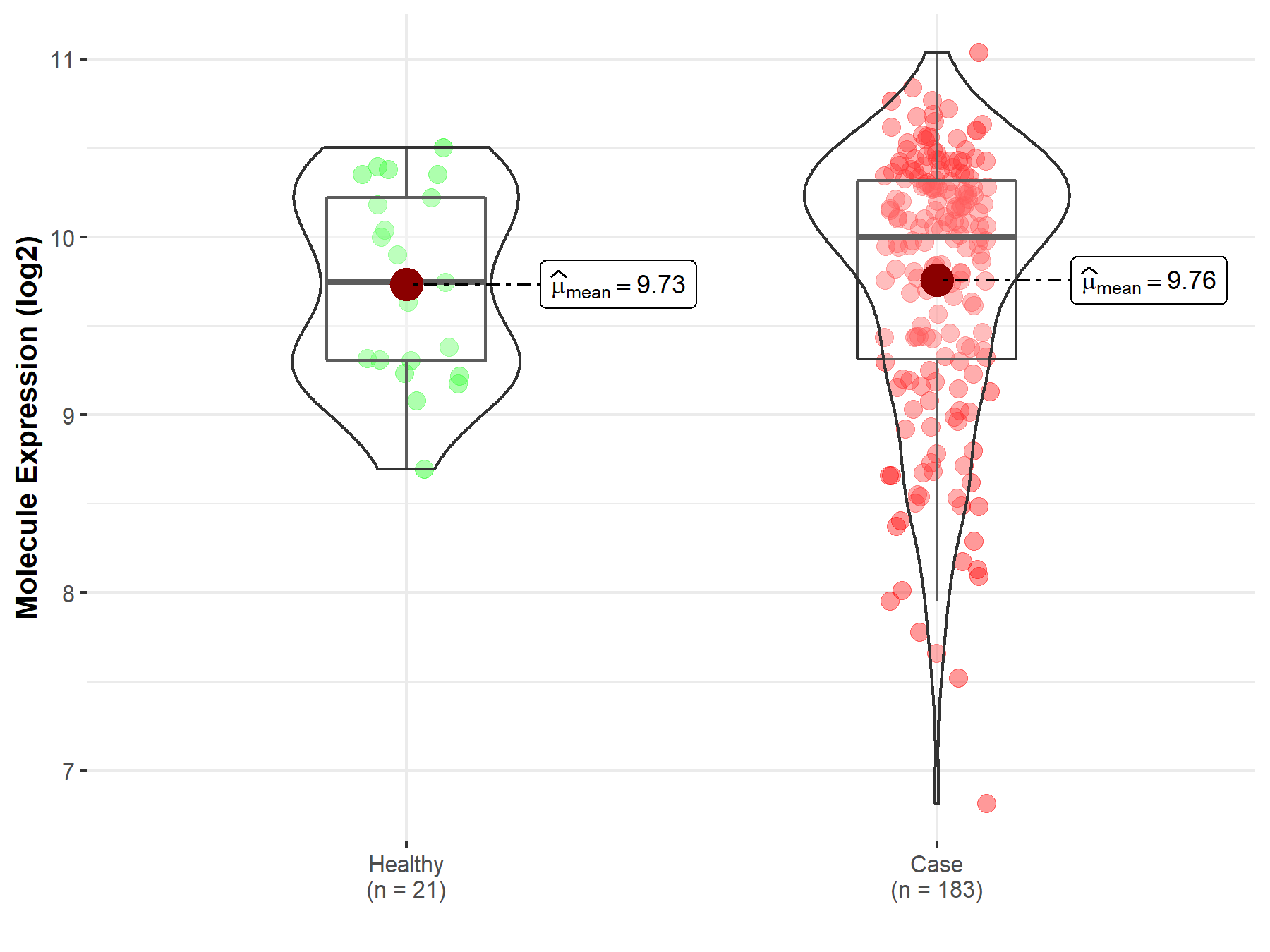
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

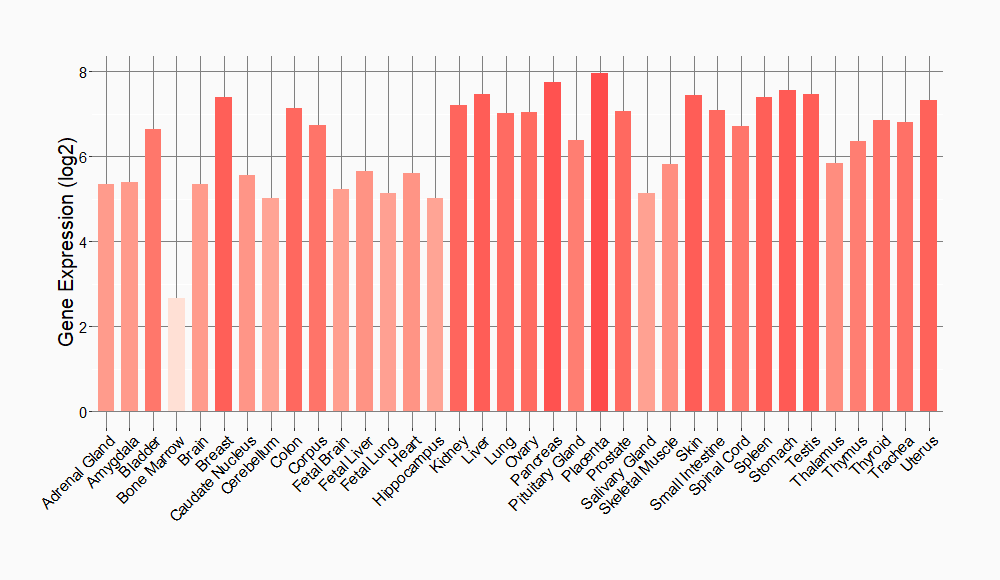
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
