Molecule Information
General Information of the Molecule (ID: Mol00711)
| Name |
DNA repair protein XRCC3 (XRCC3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
X-ray repair cross-complementing protein 3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
XRCC3
|
||||
| Gene ID | |||||
| Location |
chr14:103697609-103715504[-]
|
||||
| Sequence |
MDLDLLDLNPRIIAAIKKAKLKSVKEVLHFSGPDLKRLTNLSSPEVWHLLRTASLHLRGS
SILTALQLHQQKERFPTQHQRLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQL CLAVQFPRQHGGLEAGAVYICTEDAFPHKRLQQLMAQQPRLRTDVPGELLQKLRFGSQIF IEHVADVDTLLECVNKKVPVLLSRGMARLVVIDSVAAPFRCEFDSQASAPRARHLQSLGA TLRELSSAFQSPVLCINQVTEAMEEQGAAHGPLGFWDERVSPALGITWANQLLVRLLADR LREEEAALGCPARTLRVLSAPHLPPSSCSYTISAEGVRGTPGTQSH Click to Show/Hide
|
||||
| Function |
Involved in the homologous recombination repair (HRR) pathway of double-stranded DNA, thought to repair chromosomal fragmentation, translocations and deletions. Part of the RAD21 paralog protein complex CX3 which acts in the BRCA1-BRCA2-dependent HR pathway. Upon DNA damage, CX3 acts downstream of RAD51 recruitment; the complex binds predominantly to the intersection of the four duplex arms of the Holliday junction (HJ) and to junctions of replication forks. Involved in HJ resolution and thus in processing HR intermediates late in the DNA repair process; the function may be linked to the CX3 complex and seems to involve GEN1 during mitotic cell cycle progression. Part of a PALB2-scaffolded HR complex containing BRCA2 and RAD51C and which is thought to play a role in DNA repair by HR. Plays a role in regulating mitochondrial DNA copy number under conditions of oxidative stress in the presence of RAD51 and RAD51C.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Investigative Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Esophageal cancer [ICD-11: 2B70.1] | [1] | |||
| Sensitive Disease | Esophageal cancer [ICD-11: 2B70.1] | |||
| Sensitive Drug | 2-Bromo-4-fluorobenzaldehyde | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| EC9706 cells | Esophagus | Homo sapiens (Human) | CVCL_E307 | |
| KYSE150 cells | Esophagus | Homo sapiens (Human) | CVCL_1348 | |
| EC109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 | |
| EC1 cells | Esophagus | Homo sapiens (Human) | CVCL_DC74 | |
| HEEC cells | Esophagus | Homo sapiens (Human) | N.A. | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miroRNA-127-3p targets XRCC3 to enhance the chemosensitivity of esophageal cancer cells to a novel phenanthroline-dione derivative. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Esophagus | |
| The Specified Disease | Esophageal cancer | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.22E-02; Fold-change: 3.53E-01; Z-score: 1.09E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
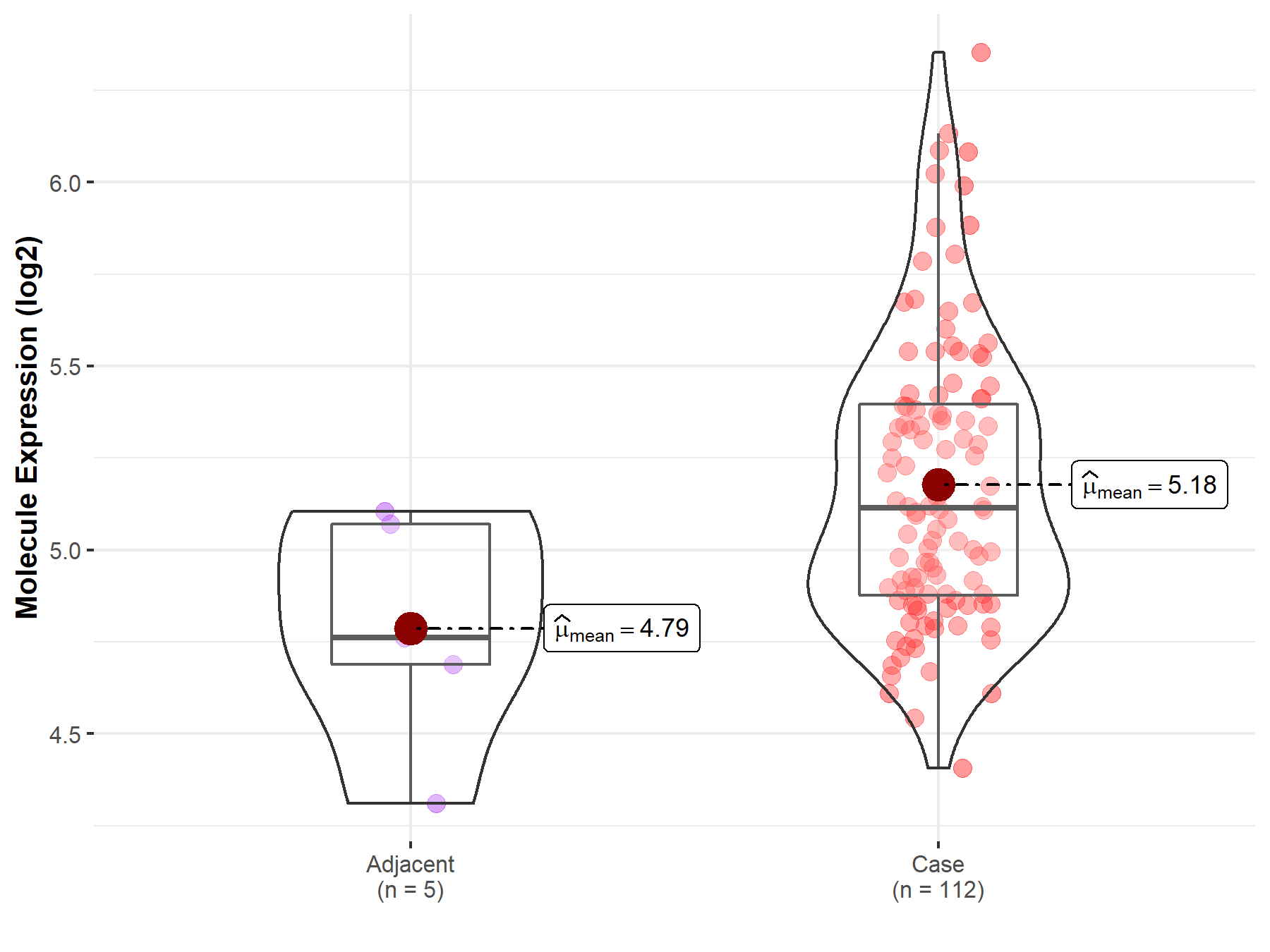
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

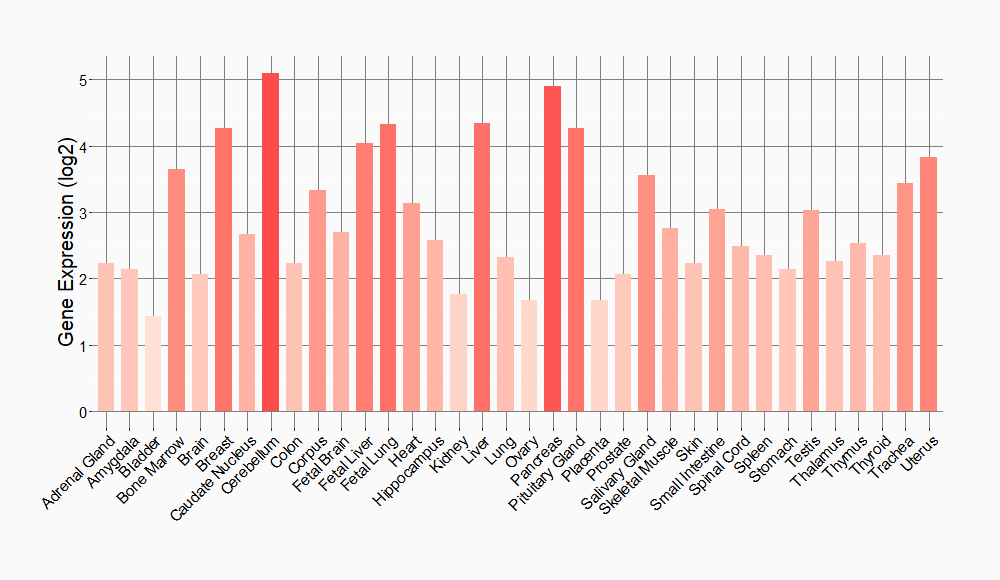
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
