Molecule Information
General Information of the Molecule (ID: Mol00696)
| Name |
Serine/threonine-protein kinase ULK1 (ULK1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Autophagy-related protein 1 homolog; ATG1; hATG1; Unc-51-like kinase 1; KIAA0722
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ULK1
|
||||
| Gene ID | |||||
| Location |
chr12:131894622-131923150[+]
|
||||
| Sequence |
MEPGRGGTETVGKFEFSRKDLIGHGAFAVVFKGRHREKHDLEVAVKCINKKNLAKSQTLL
GKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQ QIAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAAT LCGSPMYMAPEVIMSQHYDGKADLWSIGTIVYQCLTGKAPFQASSPQDLRLFYEKNKTLV PTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGS GSSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMV PAQFPGDLVAEAPSAKPPPDSLMCSGSSLVASAGLESHGRTPSPSPPCSSSPSPSGRAGP FSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSP PAHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSA PEHSPRTSGLGCRLHSAPNLSDLHVVRPKLPKPPTDPLGAVFSPPQASPPQPSHGLQSCR NLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRT LPDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESL QEKPMEIAPSAGFGGSLHPGARAGGTSSPSPVVFTVGSPPSGSTPPQGPRTRMFSAGPTG SASSSARHLVPGPCSEAPAPELPAPGHGCSFADPITANLEGAVTFEAPDLPEETLMEQEH TEILRGLRFTLLFVQHVLEIAALKGSASEAAGGPEYQLQESVVADQISLLSREWGFAEQL VLYLKVAELLSSGLQSAIDQIRAGKLCLSSTVKQVVRRLNELYKASVVSCQGLSLRLQRF FLDKQRLLDRIHSITAERLIFSHAVQMVQSAALDEMFQHREGCVPRYHKALLLLEGLQHM LSDQADIENVTKCKLCIERRLSALLTGICA Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Serine/threonine-protein kinase involved in autophagy in response to starvation. Acts upstream of phosphatidylinositol 3-kinase PIK3C3 to regulate the formation of autophagophores, the precursors of autophagosomes. Part of regulatory feedback loops in autophagy: acts both as a downstream effector and negative regulator of mammalian target of rapamycin complex 1 (mTORC1) via interaction with RPTOR. Activated via phosphorylation by AMPK and also acts as a regulator of AMPK by mediating phosphorylation of AMPK subunits PRKAA1, PRKAB2 and PRKAG1, leading to negatively regulate AMPK activity. May phosphorylate ATG13/KIAA0652 and RPTOR; however such data need additional evidences. Plays a role early in neuronal differentiation and is required for granule cell axon formation. May also phosphorylate SESN2 and SQSTM1 to regulate autophagy. Phosphorylates FLCN, promoting autophagy. Phosphorylates AMBRA1 in response to autophagy induction, releasing AMBRA1 from the cytoskeletal docking site to induce autophagosome nucleation. Phosphorylates ATG4B, leading to inhibit autophagy by decreasing both proteolytic activation and delipidation activities of ATG4B.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Dasatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.52E-01 Fold-change: -2.18E-02 Z-score: -4.52E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR106a/ULk1 signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Resazurin conversion assay | |||
| Mechanism Description | Src inhibition results in autophagy activation in NSCLC cell lines. Combining Src with autophagy inhibition results in significant cell death. Induction of ULk1 upon Scr inhibition allows for autophagy activation. Src inhibition causes induction of the ULk1 targeting microRNA-106a. Expression of the "oncogenic" miR-106a sensitizes NSCLC cells to Src inhibition. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [2] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.08E-01 Fold-change: -9.36E-03 Z-score: -6.67E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell autophagy | Inhibition | hsa04140 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR26a/b can promote apoptosis and sensitize HCC to chemotherapy via suppressing the expression of autophagy initiator ULk. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 acts as a therapeutic sensitizer in breast cancer cells by inhibiting doxorubicin-induced cytoprotective autophagy and directly targeting LAPTM4B. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [3] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Crizotinib | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| ULk1 signaling pathway | Inhibition | hsa04211 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| In Vivo Model | Nude mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULk1. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Saracatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.52E-01 Fold-change: -2.18E-02 Z-score: -4.52E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR106a/ULk1 signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Resazurin conversion assay | |||
| Mechanism Description | Src inhibition results in autophagy activation in NSCLC cell lines. Combining Src with autophagy inhibition results in significant cell death. Induction of ULk1 upon Scr inhibition allows for autophagy activation. Src inhibition causes induction of the ULk1 targeting microRNA-106a. Expression of the "oncogenic" miR-106a sensitizes NSCLC cells to Src inhibition. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.08E-01; Fold-change: -9.51E-02; Z-score: -2.15E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.07E-01; Fold-change: -1.05E-02; Z-score: -2.12E-02 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 6.43E-01; Fold-change: -9.98E-02; Z-score: -2.64E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
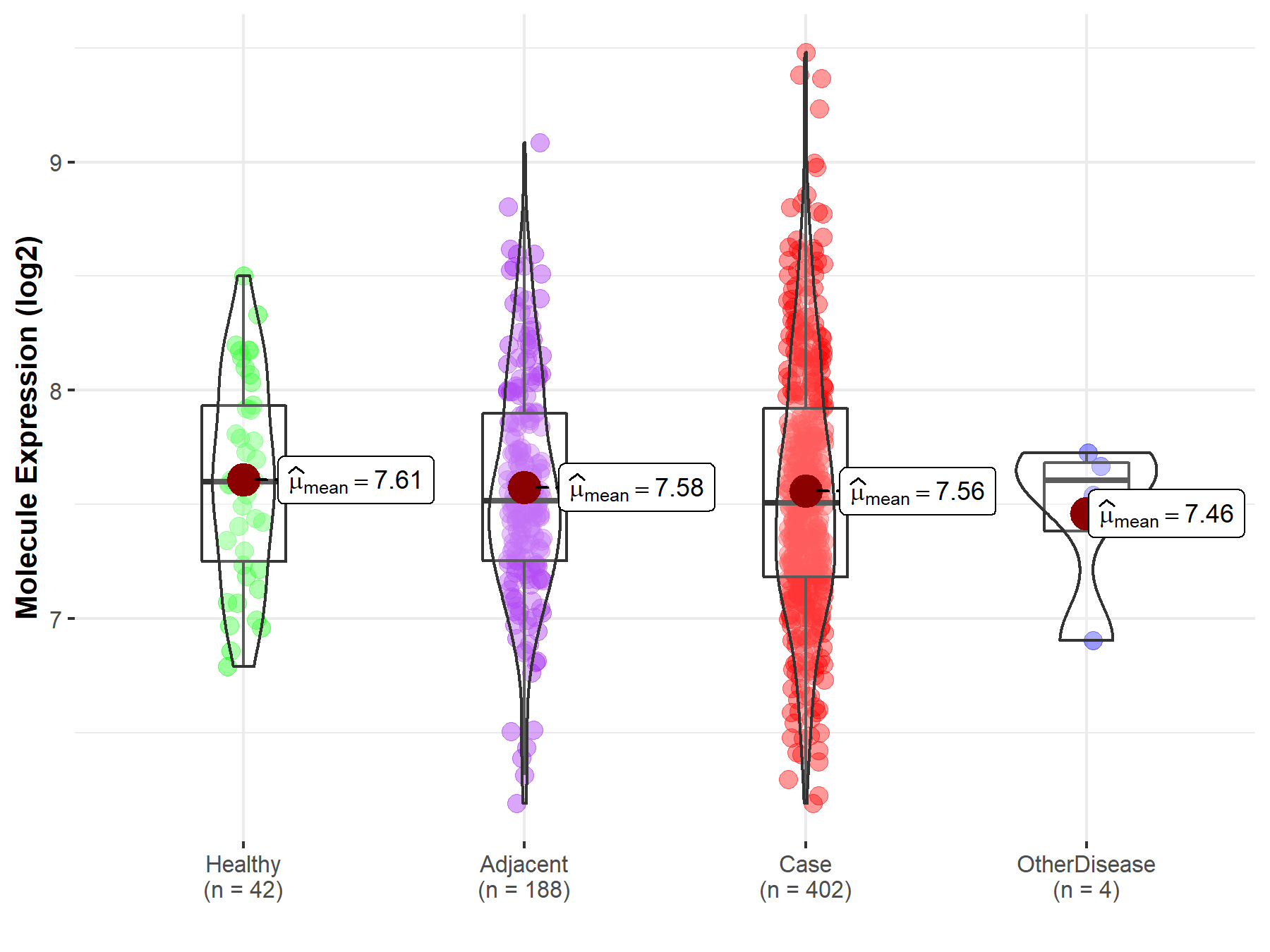
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.05E-21; Fold-change: 2.27E-01; Z-score: 8.11E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.31E-01; Fold-change: -1.82E-02; Z-score: -4.49E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.43E-41; Fold-change: 4.79E-01; Z-score: 1.03E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.13E-03; Fold-change: 3.36E-01; Z-score: 6.02E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

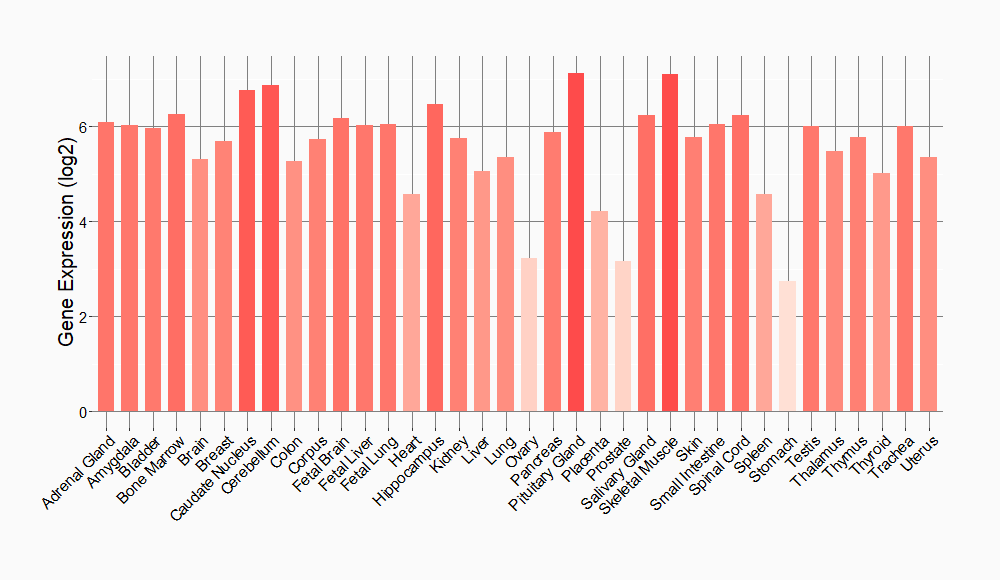
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
