Molecule Information
General Information of the Molecule (ID: Mol00677)
| Name |
Tumor necrosis factor alpha-induced protein 3 (TNFAIP3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
TNF alpha-induced protein 3; OTU domain-containing protein 7C; Putative DNA-binding protein A20; Zinc finger protein A20; OTUD7C
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
TNFAIP3
|
||||
| Gene ID | |||||
| Location |
chr6:137867214-137883312[+]
|
||||
| Sequence |
MAEQVLPQALYLSNMRKAVKIRERTPEDIFKPTNGIIHHFKTMHRYTLEMFRTCQFCPQF
REIIHKALIDRNIQATLESQKKLNWCREVRKLVALKTNGDGNCLMHATSQYMWGVQDTDL VLRKALFSTLKETDTRNFKFRWQLESLKSQEFVETGLCYDTRNWNDEWDNLIKMASTDTP MARSGLQYNSLEEIHIFVLCNILRRPIIVISDKMLRSLESGSNFAPLKVGGIYLPLHWPA QECYRYPIVLGYDSHHFVPLVTLKDSGPEIRAVPLVNRDRGRFEDLKVHFLTDPENEMKE KLLKEYLMVIEIPVQGWDHGTTHLINAAKLDEANLPKEINLVDDYFELVQHEYKKWQENS EQGRREGHAQNPMEPSVPQLSLMDVKCETPNCPFFMSVNTQPLCHECSERRQKNQNKLPK LNSKPGPEGLPGMALGASRGEAYEPLAWNPEESTGGPHSAPPTAPSPFLFSETTAMKCRS PGCPFTLNVQHNGFCERCHNARQLHASHAPDHTRHLDPGKCQACLQDVTRTFNGICSTCF KRTTAEASSSLSTSLPPSCHQRSKSDPSRLVRSPSPHSCHRAGNDAPAGCLSQAARTPGD RTGTSKCRKAGCVYFGTPENKGFCTLCFIEYRENKHFAAASGKVSPTASRFQNTIPCLGR ECGTLGSTMFEGYCQKCFIEAQNQRFHEAKRTEEQLRSSQRRDVPRTTQSTSRPKCARAS CKNILACRSEELCMECQHPNQRMGPGAHRGEPAPEDPPKQRCRAPACDHFGNAKCNGYCN ECFQFKQMYG Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Ubiquitin-editing enzyme that contains both ubiquitin ligase and deubiquitinase activities. Involved in immune and inflammatory responses signaled by cytokines, such as TNF-alpha and IL-1 beta, or pathogens via Toll-like receptors (TLRs) through terminating NF-kappa-B activity. Essential component of a ubiquitin-editing protein complex, comprising also RNF11, ITCH and TAX1BP1, that ensures the transient nature of inflammatory signaling pathways. In cooperation with TAX1BP1 promotes disassembly of E2-E3 ubiquitin protein ligase complexes in IL-1R and TNFR-1 pathways; affected are at least E3 ligases TRAF6, TRAF2 and BIRC2, and E2 ubiquitin-conjugating enzymes UBE2N and UBE2D3. In cooperation with TAX1BP1 promotes ubiquitination of UBE2N and proteasomal degradation of UBE2N and UBE2D3. Upon TNF stimulation, deubiquitinates 'Lys-63'-polyubiquitin chains on RIPK1 and catalyzes the formation of 'Lys-48'-polyubiquitin chains. This leads to RIPK1 proteasomal degradation and consequently termination of the TNF- or LPS-mediated activation of NF-kappa-B. Deubiquitinates TRAF6 probably acting on 'Lys-63'-linked polyubiquitin. Upon T-cell receptor (TCR)-mediated T-cell activation, deubiquitinates 'Lys-63'-polyubiquitin chains on MALT1 thereby mediating disassociation of the CBM (CARD11:BCL10:MALT1) and IKK complexes and preventing sustained IKK activation. Deubiquitinates NEMO/IKBKG; the function is facilitated by TNIP1 and leads to inhibition of NF-kappa-B activation. Upon stimulation by bacterial peptidoglycans, probably deubiquitinates RIPK2. Can also inhibit I-kappa-B-kinase (IKK) through a non-catalytic mechanism which involves polyubiquitin; polyubiquitin promotes association with IKBKG and prevents IKK MAP3K7-mediated phosphorylation. Targets TRAF2 for lysosomal degradation. In vitro able to deubiquitinate 'Lys-11'-, 'Lys-48'- and 'Lys-63' polyubiquitin chains. Inhibitor of programmed cell death. Has a role in the function of the lymphoid system. Required for LPS-induced production of proinflammatory cytokines and IFN beta in LPS-tolerized macrophages.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [1] | |||
| Resistant Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Resistant Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.21E-01 Fold-change: -4.55E-03 Z-score: -1.00E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| SW1990 cells | Pancreas | Homo sapiens (Human) | CVCL_1723 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-125a may promote chemo-resistance to gemcitabine in pancreatic cell lines through targeting A20, which may provide novel therapeutic targets or molecular biomarkers for cancer therapy and improve tumor diagnosis or predictions of therapeutic responses. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [2] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Neuroectodermal tumor | |||
| The Studied Tissue | Brainstem tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.91E-03 Fold-change: -2.09E-01 Z-score: -3.84E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| LN-18 cells | Brain | Homo sapiens (Human) | CVCL_0392 | |
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| HS683 cells | Brain | Homo sapiens (Human) | CVCL_0844 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | A novel mechanism independent of TP53 and MGMT by which oncogenic miR-125b confers TMZ resistance by targeting TNFAIP3 and NkIRAS2. GBM cells overexpressing miR-125b showed increased NF-kB activity and upregulation of anti-apoptotic and cell cycle genes. This was significantly associated with resistance of GBM cells to TNFalpha- and TNF-related inducing ligand-induced apoptosis as well as resistance to TMZ. Conversely, overexpression of anti-miR-125b resulted in cell cycle arrest, increased apoptosis and increased sensitivity to TMZ, indicating that endogenous miR-125b is sufficient to control these processes. GBM cells overexpressing TNFAIP3 and NkIRAS2 were refractory to miR-125b-induced apoptosis resistance as well as TMZ resistance, indicating that both genes are relevant targets of miR-125b. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.49E-91; Fold-change: 1.13E+00; Z-score: 1.54E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
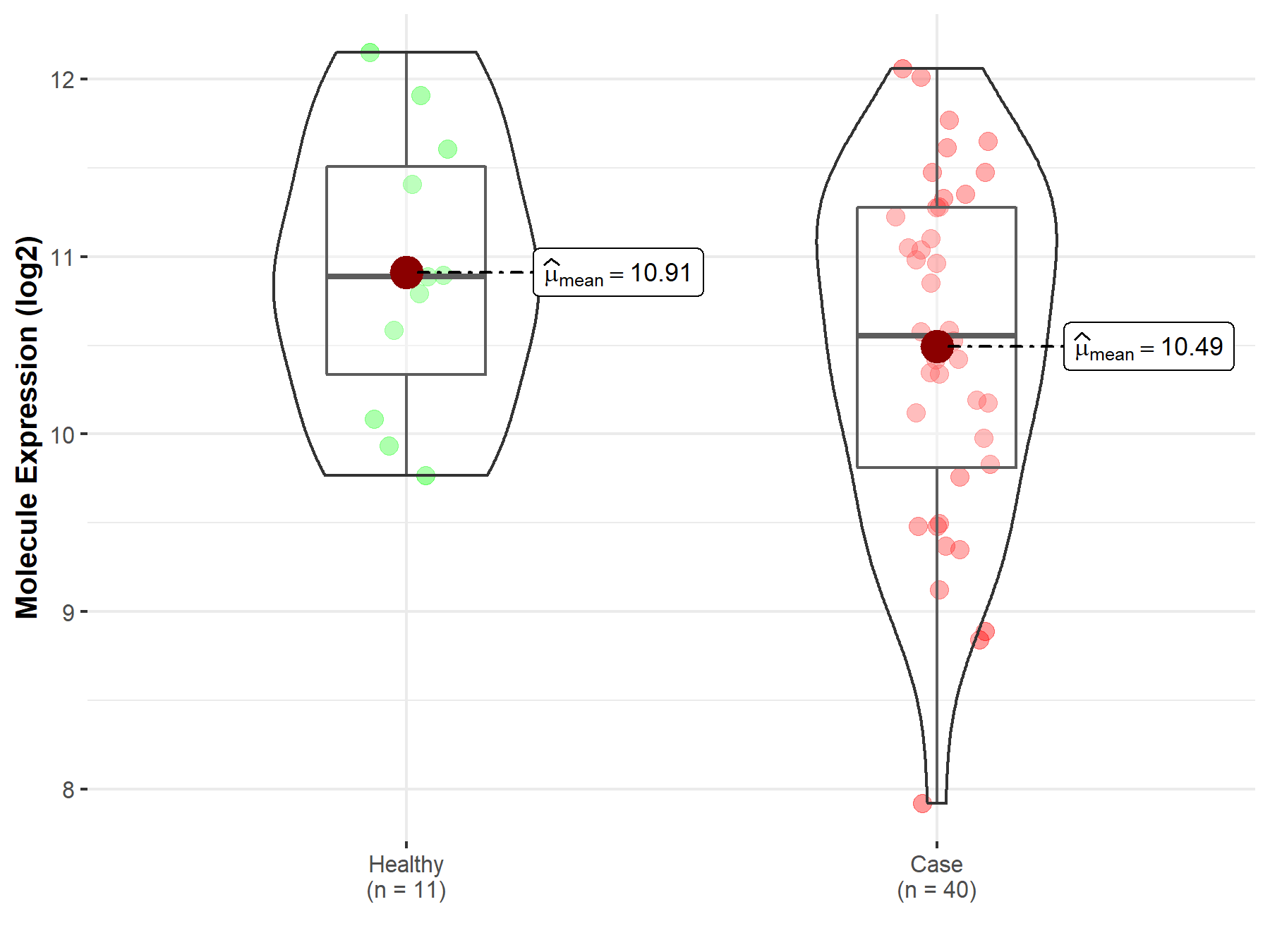
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.73E-01; Fold-change: -9.58E-02; Z-score: -5.36E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
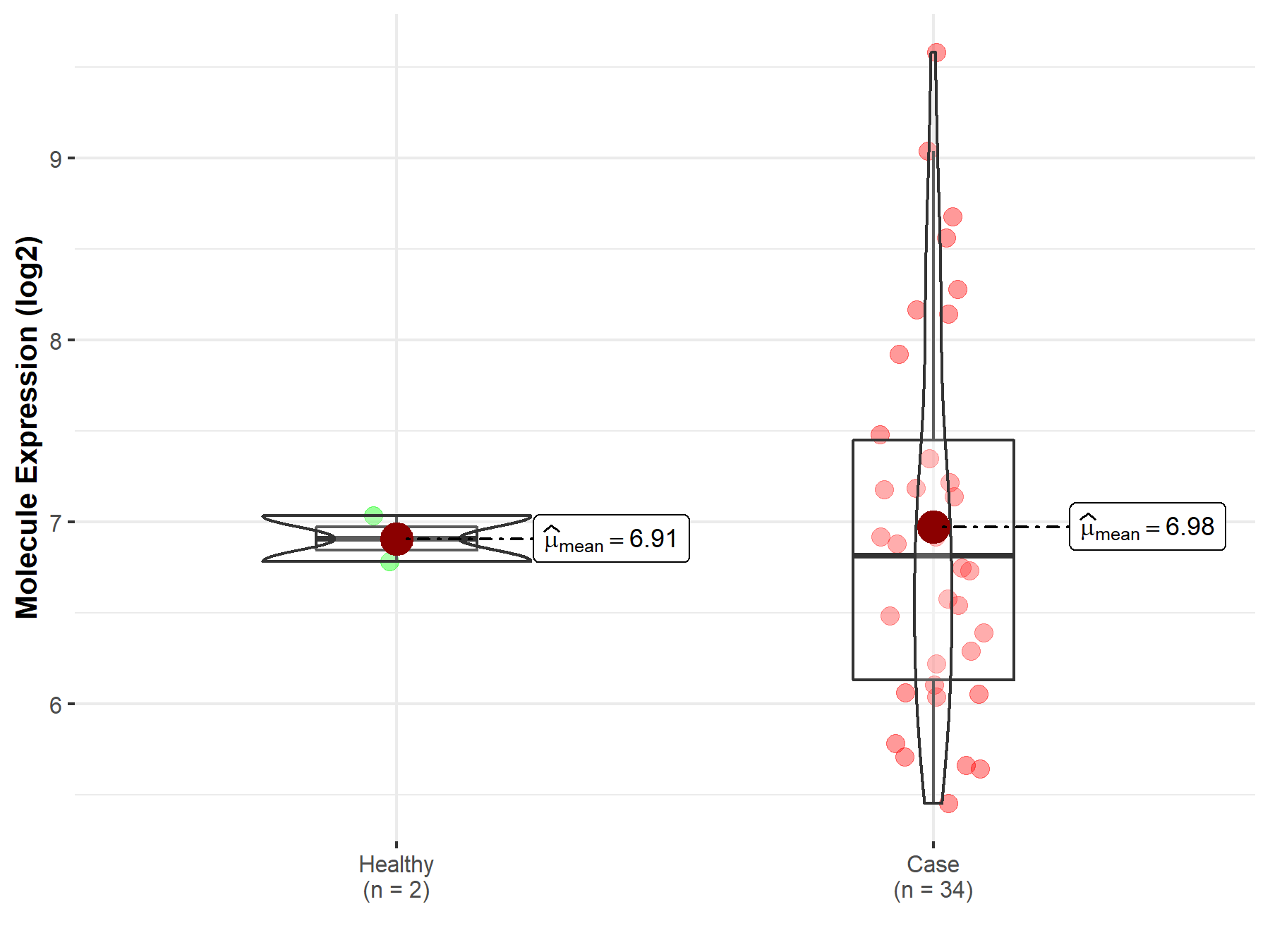
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-01; Fold-change: -3.37E-01; Z-score: -4.23E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.91E-03; Fold-change: -1.07E+00; Z-score: -1.35E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.21E-01; Fold-change: 2.88E-01; Z-score: 2.62E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.89E-02; Fold-change: 6.15E-01; Z-score: 4.46E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
