Molecule Information
General Information of the Molecule (ID: Mol00636)
| Name |
SWI/SNF complex subunit SMARCC1 (SMARCC1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
BRG1-associated factor 155; BAF155; SWI/SNF complex 155 kDa subunit; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1; BAF155
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
SMARCC1
|
||||
| Gene ID | |||||
| Location |
chr3:47585269-47782106[-]
|
||||
| Sequence |
MAAAAGGGGPGTAVGATGSGIAAAAAGLAVYRRKDGGPATKFWESPETVSQLDSVRVWLG
KHYKKYVHADAPTNKTLAGLVVQLLQFQEDAFGKHVTNPAFTKLPAKCFMDFKAGGALCH ILGAAYKYKNEQGWRRFDLQNPSRMDRNVEMFMNIEKTLVQNNCLTRPNIYLIPDIDLKL ANKLKDIIKRHQGTFTDEKSKASHHIYPYSSSQDDEEWLRPVMRKEKQVLVHWGFYPDSY DTWVHSNDVDAEIEDPPIPEKPWKVHVKWILDTDIFNEWMNEEDYEVDENRKPVSFRQRI STKNEEPVRSPERRDRKASANARKRKHSPSPPPPTPTESRKKSGKKGQASLYGKRRSQKE EDEQEDLTKDMEDPTPVPNIEEVVLPKNVNLKKDSENTPVKGGTVADLDEQDEETVTAGG KEDEDPAKGDQSRSVDLGEDNVTEQTNHIIIPSYASWFDYNCIHVIERRALPEFFNGKNK SKTPEIYLAYRNFMIDTYRLNPQEYLTSTACRRNLTGDVCAVMRVHAFLEQWGLVNYQVD PESRPMAMGPPPTPHFNVLADTPSGLVPLHLRSPQVPAAQQMLNFPEKNKEKPVDLQNFG LRTDIYSKKTLAKSKGASAGREWTEQETLLLLEALEMYKDDWNKVSEHVGSRTQDECILH FLRLPIEDPYLENSDASLGPLAYQPVPFSQSGNPVMSTVAFLASVVDPRVASAAAKAALE EFSRVREEVPLELVEAHVKKVQEAARASGKVDPTYGLESSCIAGTGPDEPEKLEGAEEEK MEADPDGQQPEKAENKVENETDEGDKAQDGENEKNSEKEQDSEVSEDTKSEEKETEENKE LTDTCKERESDTGKKKVEHEISEGNVATAAAAALASAATKAKHLAAVEERKIKSLVALLV ETQMKKLEIKLRHFEELETIMDREKEALEQQRQQLLTERQNFHMEQLKYAELRARQQMEQ QQHGQNPQQAHQHSGGPGLAPLGAAGHPGMMPHQQPPPYPLMHHQMPPPHPPQPGQIPGP GSMMPGQHMPGRMIPTVAANIHPSGSGPTPPGMPPMPGNILGPRVPLTAPNGMYPPPPQQ QPPPPPPADGVPPPPAPGPPASAAP Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. May stimulate the ATPase activity of the catalytic subunit of the complex. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [1] | |||
| Resistant Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Resistant Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.52E-01 Fold-change: -1.99E-03 Z-score: -6.16E-02 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 |
| PSN1 cells | Pancreas | Homo sapiens (Human) | CVCL_1644 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-320c regulates the resistance of pancreatic cancer cells to gemcitabine through SMARCC1. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.52E-01; Fold-change: -1.95E-01; Z-score: -2.91E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.13E-20; Fold-change: 4.22E-01; Z-score: 1.60E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
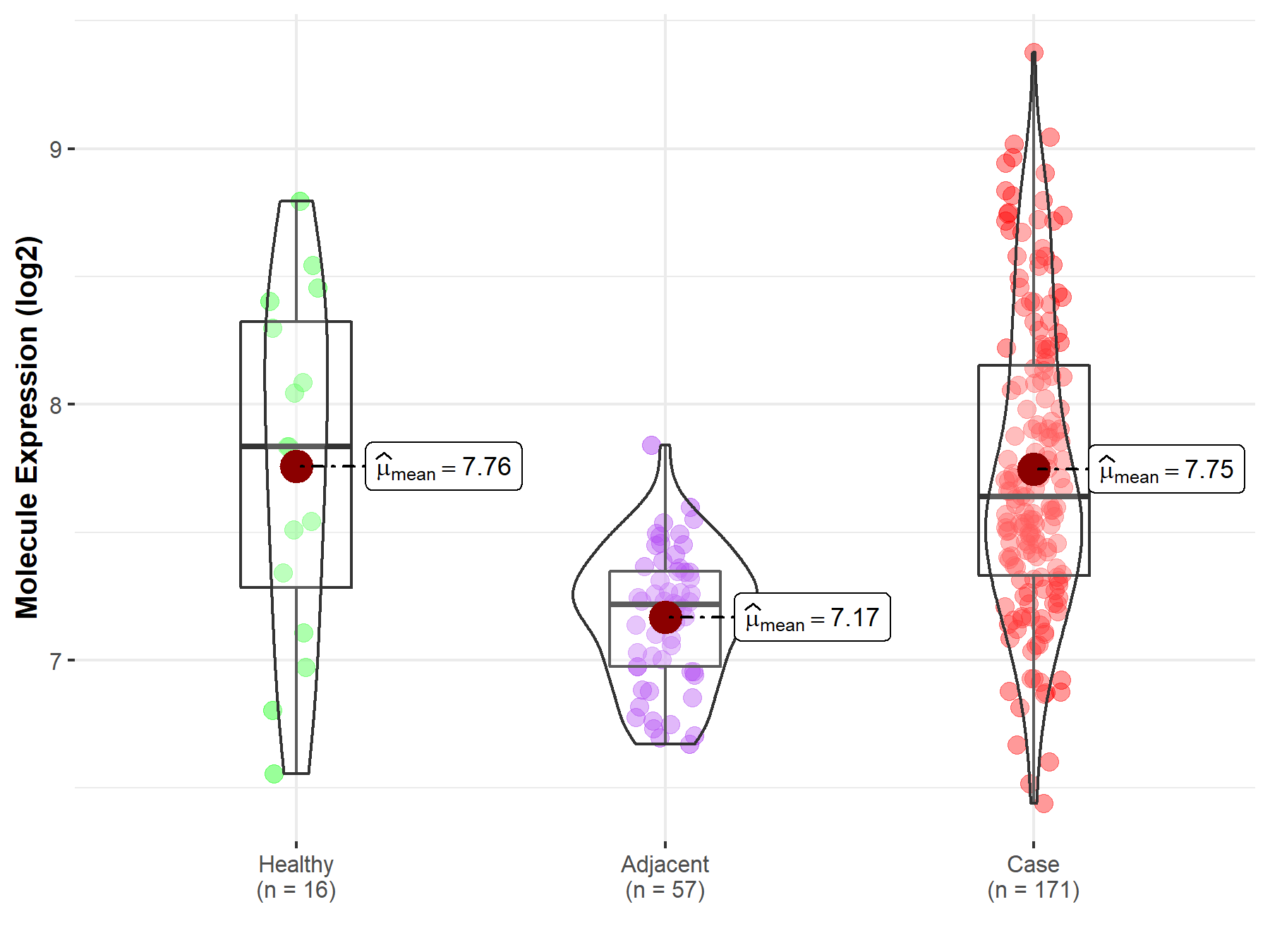
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
