Molecule Information
General Information of the Molecule (ID: Mol00608)
| Name |
hnRNP A2/B1 (HNRNPA2B1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
hnRNP A2/B1; HNRPA2B1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
HNRNPA2B1
|
||||
| Gene ID | |||||
| Location |
chr7:26171151-26201529[-]
|
||||
| Sequence |
MEKTLETVPLERKKREKEQFRKLFIGGLSFETTEESLRNYYEQWGKLTDCVVMRDPASKR
SRGFGFVTFSSMAEVDAAMAARPHSIDGRVVEPKRAVAREESGKPGAHVTVKKLFVGGIK EDTEEHHLRDYFEEYGKIDTIEIITDRQSGKKRGFGFVTFDDHDPVDKIVLQKYHTINGH NAEVRKALSRQEMQEVQSSRSGRGGNFGFGDSRGGGGNFGPGPGSNFRGGSDGYGSGRGF GDGYNGYGGGPGGGNFGGSPGYGGGRGGYGGGGPGYGNQGGGYGGGYDNYGGGNYGSGNY NDFGNYNQQPSNYGPMKSGNFGGSRNMGGPYGGGNYGPGGSGGSGGYGGRSRY Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Heterogeneous nuclear ribonucleoprotein (hnRNP) that associates with nascent pre-mRNAs, packaging them into hnRNP particles. The hnRNP particle arrangement on nascent hnRNA is non-random and sequence-dependent and serves to condense and stabilize the transcripts and minimize tangling and knotting. Packaging plays a role in various processes such as transcription, pre-mRNA processing, RNA nuclear export, subcellular location, mRNA translation and stability of mature mRNAs. Forms hnRNP particles with at least 20 other different hnRNP and heterogeneous nuclear RNA in the nucleus. Involved in transport of specific mRNAs to the cytoplasm in oligodendrocytes and neurons: acts by specifically recognizing and binding the A2RE (21 nucleotide hnRNP A2 response element) or the A2RE11 (derivative 11 nucleotide oligonucleotide) sequence motifs present on some mRNAs, and promotes their transport to the cytoplasm. Specifically binds single-stranded telomeric DNA sequences, protecting telomeric DNA repeat against endonuclease digestion. Also binds other RNA molecules, such as primary miRNA (pri-miRNAs): acts as a nuclear 'reader' of the N6-methyladenosine (m6A) mark by specifically recognizing and binding a subset of nuclear m6A-containing pri-miRNAs. Binding to m6A-containing pri-miRNAs promotes pri-miRNA processing by enhancing binding of DGCR8 to pri-miRNA transcripts. Involved in miRNA sorting into exosomes following sumoylation, possibly by binding (m6A)-containing pre-miRNAs. Acts as a regulator of efficiency of mRNA splicing, possibly by binding to m6A-containing pre-mRNAs. Plays a role in the splicing of pyruvate kinase PKM by binding repressively to sequences flanking PKM exon 9, inhibiting exon 9 inclusion and resulting in exon 10 inclusion and production of the PKM M2 isoform. Also plays a role in the activation of the innate immune response. Mechanistically, senses the presence of viral DNA in the nucleus, homodimerizes and is demethylated by JMJD6. In turn, translocates to the cytoplasm where it activates the TBK1-IRF3 pathway, leading to interferon alpha/beta production.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [1] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 |
| HCC4006 cells | Lung | Homo sapiens (Human) | CVCL_1269 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Knockdown of H19 by siRNA transfection can significantly reduce the expression of N-cadherin, as well as increase E-cadherin and vimentin level, which improved tamoxifen sensitivity in tamoxifen-resistant breast cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | Exosomal AGAP2-AS1 expression was upregulated by hnRNPA2B1 overexpression and suppressed by hnRNPA2B1 knockdown in SkBR-3R cells while knockdown of AGAP2-AS1 resensitized trastuzumab resistance in breast cancer cells. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.94E-05; Fold-change: 2.94E-01; Z-score: 4.27E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.15E-03; Fold-change: 1.58E-01; Z-score: 1.61E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.28E-27; Fold-change: 1.00E+00; Z-score: 1.14E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.39E-02; Fold-change: 3.26E-01; Z-score: 4.74E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
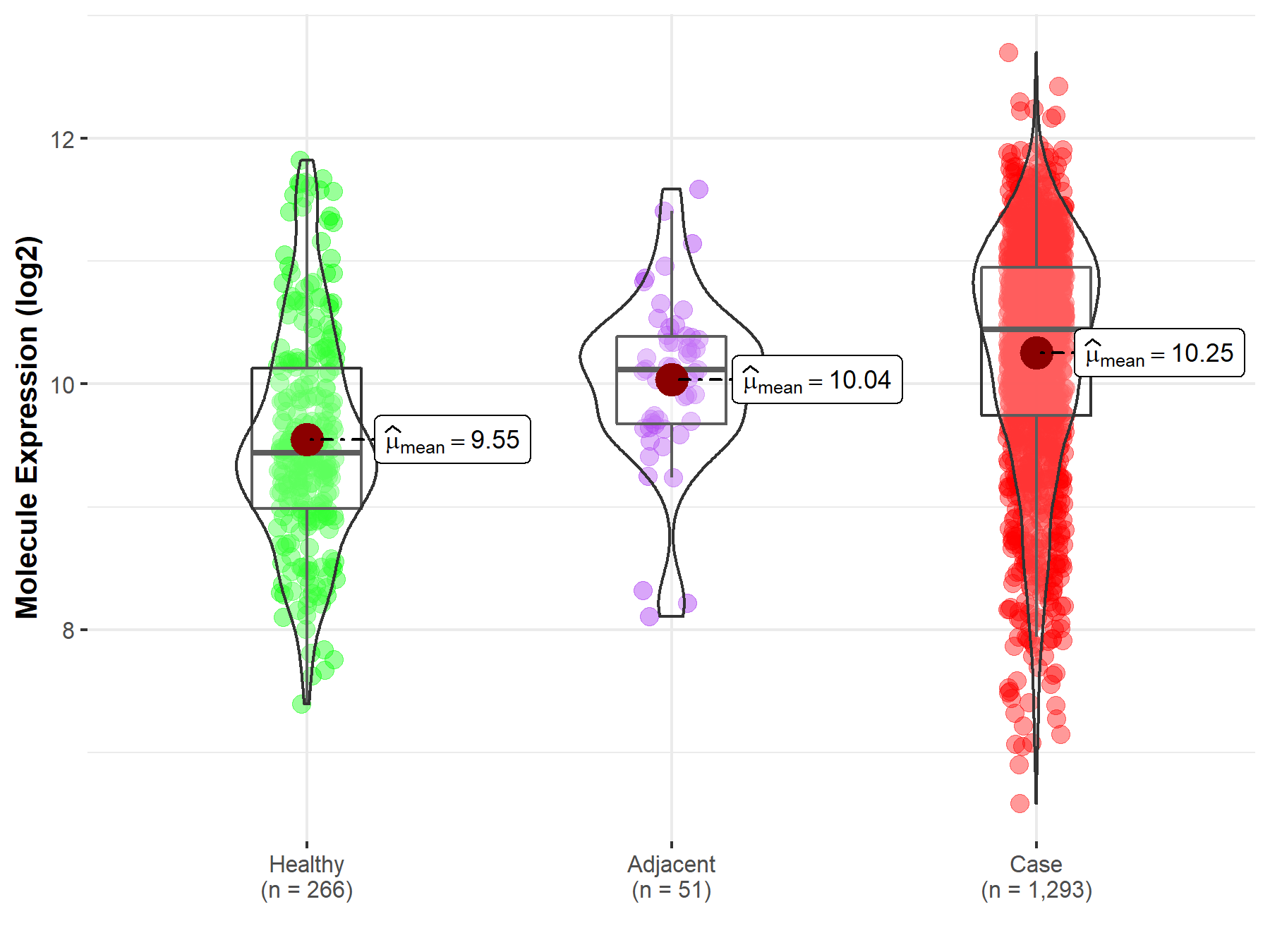
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
