Molecule Information
General Information of the Molecule (ID: Mol00594)
| Name |
Retinoblastoma-associated protein (RB1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
p105-Rb; p110-RB1; pRb; Rb; pp110
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
RB1
|
||||
| Gene ID | |||||
| Location |
chr13:48303744-48599436[+]
|
||||
| Sequence |
MPPKTPRKTAATAAAAAAEPPAPPPPPPPEEDPEQDSGPEDLPLVRLEFEETEEPDFTAL
CQKLKIPDHVRERAWLTWEKVSSVDGVLGGYIQKKKELWGICIFIAAVDLDEMSFTFTEL QKNIEISVHKFFNLLKEIDTSTKVDNAMSRLLKKYDVLFALFSKLERTCELIYLTQPSSS ISTEINSALVLKVSWITFLLAKGEVLQMEDDLVISFQLMLCVLDYFIKLSPPMLLKEPYK TAVIPINGSPRTPRRGQNRSARIAKQLENDTRIIEVLCKEHECNIDEVKNVYFKNFIPFM NSLGLVTSNGLPEVENLSKRYEEIYLKNKDLDARLFLDHDKTLQTDSIDSFETQRTPRKS NLDEEVNVIPPHTPVRTVMNTIQQLMMILNSASDQPSENLISYFNNCTVNPKESILKRVK DIGYIFKEKFAKAVGQGCVEIGSQRYKLGVRLYYRVMESMLKSEEERLSIQNFSKLLNDN IFHMSLLACALEVVMATYSRSTSQNLDSGTDLSFPWILNVLNLKAFDFYKVIESFIKAEG NLTREMIKHLERCEHRIMESLAWLSDSPLFDLIKQSKDREGPTDHLESACPLNLPLQNNH TAADMYLSPVRSPKKKGSTTRVNSTANAETQATSAFQTQKPLKSTSLSLFYKKVYRLAYL RLNTLCERLLSEHPELEHIIWTLFQHTLQNEYELMRDRHLDQIMMCSMYGICKVKNIDLK FKIIVTAYKDLPHAVQETFKRVLIKEEEYDSIIVFYNSVFMQRLKTNILQYASTRPPTLS PIPHIPRSPYKFPSSPLRIPGGNIYISPLKSPYKISEGLPTPTKMTPRSRILVSIGESFG TSEKFQKINQMVCNSDRVLKRSAEGSNPPKPLKKLRFDIEGSDEADGSKHLPGESKFQQK LAEMTSTRTRMQKQKMNDSMDTSNKEEK Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Tumor suppressor that is a key regulator of the G1/S transition of the cell cycle. The hypophosphorylated form binds transcription regulators of the E2F family, preventing transcription of E2F-responsive genes. Both physically blocks E2Fs transactivating domain and recruits chromatin-modifying enzymes that actively repress transcription. Cyclin and CDK-dependent phosphorylation of RB1 induces its dissociation from E2Fs, thereby activating transcription of E2F responsive genes and triggering entry into S phase. RB1 also promotes the G0-G1 transition upon phosphorylation and activation by CDK3/cyclin-C. Directly involved in heterochromatin formation by maintaining overall chromatin structure and, in particular, that of constitutive heterochromatin by stabilizing histone methylation. Recruits and targets histone methyltransferases SUV39H1, KMT5B and KMT5C, leading to epigenetic transcriptional repression. Controls histone H4 'Lys-20' trimethylation. Inhibits the intrinsic kinase activity of TAF1. Mediates transcriptional repression by SMARCA4/BRG1 by recruiting a histone deacetylase (HDAC) complex to the c-FOS promoter. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
4 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.73E-02 Fold-change: -1.02E-02 Z-score: -1.66E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| MCF7/TAMR cells | Breast | Homo sapiens (Human) | CVCL_EG55 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| HEK293 FT cells | Kidney | Homo sapiens (Human) | CVCL_6911 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Tamoxifen-resistant cells express miRNA-519a at high levels, which directly represses the expression of PTEN, RB1, and CDkN1A, central nodes of a dense network, allowing the cells to proliferate, even in the presence of tamoxifen. miRNA-519a increases viability and S-phase population of the cell cycle, but does not affect EMT or invasion. miRNA-519a-expressing cells evade tamoxifen-induced apoptosis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [2] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Missense mutation | p.E580X |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Angiogenic potential | Inhibition | hsa04370 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [2] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Missense mutation | p.E580X |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [3] | |||
| Resistant Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| PATU8988 cells | Pancreas | Homo sapiens (Human) | CVCL_1846 | |
| 293TN cells | Pancreas | Homo sapiens (Human) | CVCL_UL49 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
5-FU and gemcitabine assay; CCK8 assay; Wound healing assay; Transwell chamber invasion assay | |||
| Mechanism Description | miRNA-221-3p desensitizes pancreatic cancer cells to 5-fluorouracil by targeting RB1. miR221-3p down-regulated RB1 expression by directly binding to its 3'-UTR and therefore caused increased several aspects of pancreatic cancer pathogenesis, including proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [4] | |||
| Resistant Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Resistant Drug | Palbociclib | |||
| Molecule Alteration | FS-insertion | p.M695fs*26 (c.2083_2084insC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| Mechanism Description | The combination of CDK4/6 and PI3K inhibition induces a different mode of arrest compared with palbociclib alone, characterized by not only sustained growth arrest, but also increased apoptosis in vitro, as well as tumor regression in vivo. | |||
Discontinued Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [5] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Rociletinib | |||
| Molecule Alteration | Single nucleotide variants | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Circulating tumour DNA (ctDNA) analysis | |||
| Experiment for Drug Resistance |
Tissue biopsy assay; CT scan assay | |||
| Mechanism Description | Rociletinib resistance recurrently involves MET, EGFR, PIk3CA, ERRB2, kRAS and RB1. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [6] | |||
| Resistant Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | |||
| Resistant Drug | Letrozole/Ribociclib | |||
| Molecule Alteration | Missense mutation | p.H483Y (c.1447C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Blood sample | N.A. | ||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.75E-02; Fold-change: 2.12E-01; Z-score: 4.97E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.50E-03; Fold-change: 2.34E-01; Z-score: 3.99E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.26E-08; Fold-change: -1.55E-01; Z-score: -4.31E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.21E-04; Fold-change: -6.24E-02; Z-score: -1.58E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.73E-02; Fold-change: -1.31E-01; Z-score: -3.14E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.57E-01; Fold-change: -6.74E-02; Z-score: -1.25E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
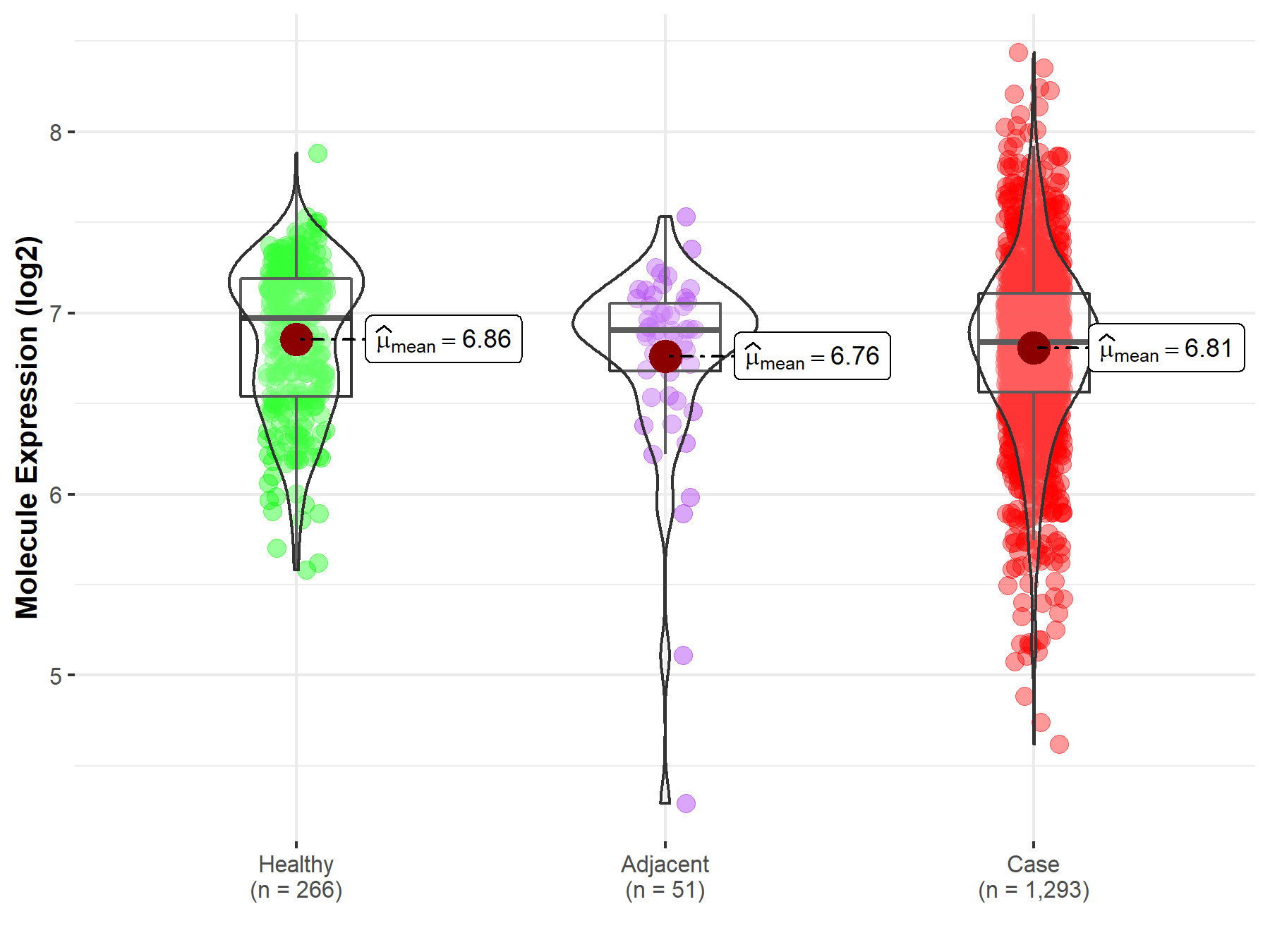
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.38E-02; Fold-change: 3.22E-01; Z-score: 6.39E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.06E-02; Fold-change: -1.66E-01; Z-score: -2.73E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

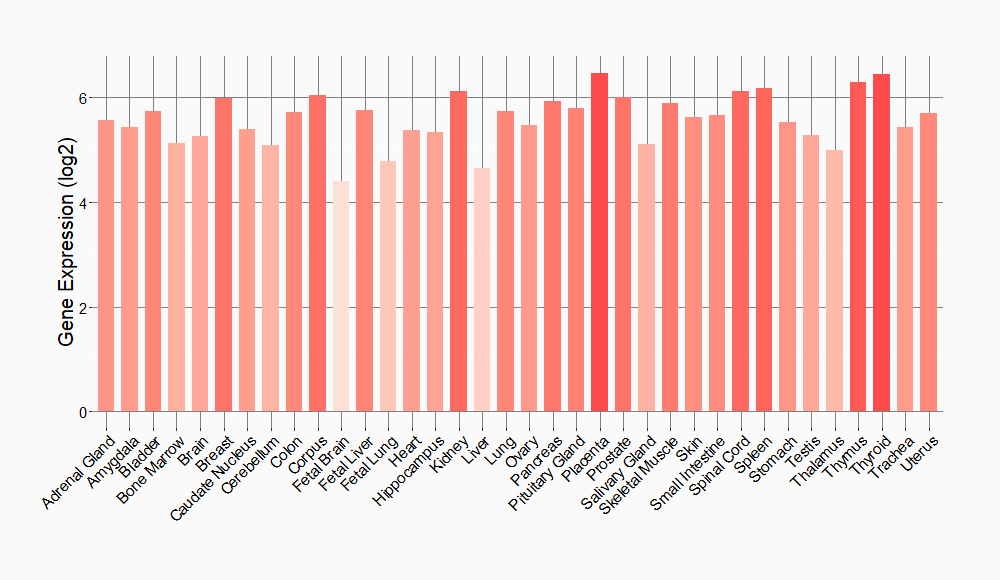
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
