Molecule Information
General Information of the Molecule (ID: Mol00572)
| Name |
Proteasome subunit beta type-5 (PSMB5)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Macropain epsilon chain; Multicatalytic endopeptidase complex epsilon chain; Proteasome chain 6; Proteasome epsilon chain; Proteasome subunit MB1; Proteasome subunit X; LMPX; MB1; X
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PSMB5
|
||||
| Gene ID | |||||
| Location |
chr14:23016543-23035230[-]
|
||||
| Sequence |
MALASVLERPLPVNQRGFFGLGGRADLLDLGPGSLSDGLSLAAPGWGVPEEPGIEMLHGT
TTLAFKFRHGVIVAADSRATAGAYIASQTVKKVIEINPYLLGTMAGGAADCSFWERLLAR QCRIYELRNKERISVAAASKLLANMVYQYKGMGLSMGTMICGWDKRGPGLYYVDSEGNRI SGATFSVGSGSVYAYGVMDRGYSYDLEVEQAYDLARRAIYQATYRDAYSGGAVNLYHVRE DGWIRVSSDNVADLHEKYSGSTP Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Component of the 20S core proteasome complex involved in the proteolytic degradation of most intracellular proteins. This complex plays numerous essential roles within the cell by associating with different regulatory particles. Associated with two 19S regulatory particles, forms the 26S proteasome and thus participates in the ATP-dependent degradation of ubiquitinated proteins. The 26S proteasome plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins that could impair cellular functions, and by removing proteins whose functions are no longer required. Associated with the PA200 or PA28, the 20S proteasome mediates ubiquitin-independent protein degradation. This type of proteolysis is required in several pathways including spermatogenesis (20S-PA200 complex) or generation of a subset of MHC class I-presented antigenic peptides (20S-PA28 complex). Within the 20S core complex, PSMB5 displays a chymotrypsin-like activity.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [1] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Bortezomib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| PI3K/RAS signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Bone marrow | Blood | Homo sapiens (Human) | N.A. |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Gene expression profiling assay; High-resolution copy number arrays assay; Whole-exome sequencing assay | |||
| Experiment for Drug Resistance |
Longitudinal copy number aberration (CNA) analysis | |||
| Mechanism Description | Resistance to immunomodulatory drugs (IMiD) and proteasome inhibitors was recently associated with mutations in IMiD response genes IRF4, CRBN, DDB1, CUL4A, CUL4B, IkZF1, IkZF2, and IkZF3 or in the proteasome inhibitor response genes PSMB5 and PSMG2, respectively. Mechanistically, bi-allelic loss of tumor-suppressor genes is a crucial mechanism, allowing units of selection to evade treatment-induced apoptosis with the acquisition of subsequent proliferative advantage leading to their outgrowth. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [1] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Cortiosteroids | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| PI3K/RAS signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Bone marrow | Blood | Homo sapiens (Human) | N.A. |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Gene expression profiling assay; High-resolution copy number arrays assay; Whole-exome sequencing assay | |||
| Experiment for Drug Resistance |
Longitudinal copy number aberration (CNA) analysis | |||
| Mechanism Description | Resistance to immunomodulatory drugs (IMiD) and proteasome inhibitors was recently associated with mutations in IMiD response genes IRF4, CRBN, DDB1, CUL4A, CUL4B, IkZF1, IkZF2, and IkZF3 or in the proteasome inhibitor response genes PSMB5 and PSMG2, respectively. Mechanistically, bi-allelic loss of tumor-suppressor genes is a crucial mechanism, allowing units of selection to evade treatment-induced apoptosis with the acquisition of subsequent proliferative advantage leading to their outgrowth. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.50E-06; Fold-change: 1.09E+00; Z-score: 4.18E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
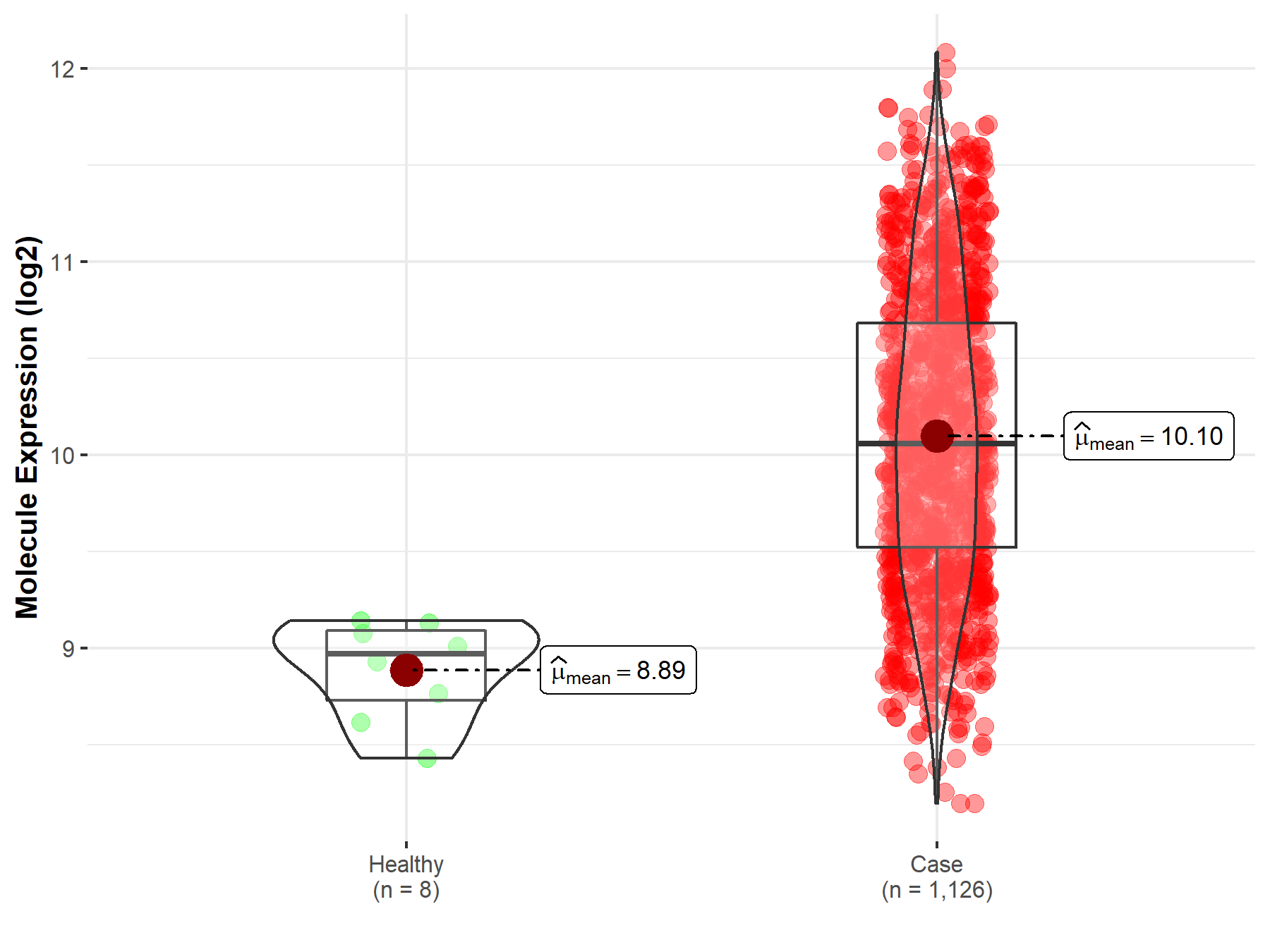
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.62E-01; Fold-change: -1.06E-01; Z-score: -1.18E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
