Molecule Information
General Information of the Molecule (ID: Mol00546)
| Name |
PI3-kinase regulatory subunit beta (PIK3R2)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
PI3-kinase regulatory subunit beta; PI3K regulatory subunit beta; PtdIns-3-kinase regulatory subunit beta; Phosphatidylinositol 3-kinase 85 kDa regulatory subunit beta; PI3-kinase subunit p85-beta; PtdIns-3-kinase regulatory subunit p85-beta
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PIK3R2
|
||||
| Gene ID | |||||
| Location |
chr19:18153163-18170532[+]
|
||||
| Sequence |
MAGPEGFQYRALYPFRRERPEDLELLPGDVLVVSRAALQALGVAEGGERCPQSVGWMPGL
NERTRQRGDFPGTYVEFLGPVALARPGPRPRGPRPLPARPRDGAPEPGLTLPDLPEQFSP PDVAPPLLVKLVEAIERTGLDSESHYRPELPAPRTDWSLSDVDQWDTAALADGIKSFLLA LPAPLVTPEASAEARRALREAAGPVGPALEPPTLPLHRALTLRFLLQHLGRVASRAPALG PAVRALGATFGPLLLRAPPPPSSPPPGGAPDGSEPSPDFPALLVEKLLQEHLEEQEVAPP ALPPKPPKAKPASTVLANGGSPPSLQDAEWYWGDISREEVNEKLRDTPDGTFLVRDASSK IQGEYTLTLRKGGNNKLIKVFHRDGHYGFSEPLTFCSVVDLINHYRHESLAQYNAKLDTR LLYPVSKYQQDQIVKEDSVEAVGAQLKVYHQQYQDKSREYDQLYEEYTRTSQELQMKRTA IEAFNETIKIFEEQGQTQEKCSKEYLERFRREGNEKEMQRILLNSERLKSRIAEIHESRT KLEQQLRAQASDNREIDKRMNSLKPDLMQLRKIRDQYLVWLTQKGARQKKINEWLGIKNE TEDQYALMEDEDDLPHHEERTWYVGKINRTQAEEMLSGKRDGTFLIRESSQRGCYACSVV VDGDTKHCVIYRTATGFGFAEPYNLYGSLKELVLHYQHASLVQHNDALTVTLAHPVRAPG PGPPPAAR Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Regulatory subunit of phosphoinositide-3-kinase (PI3K), a kinase that phosphorylates PtdIns(4,5)P2 (Phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Binds to activated (phosphorylated) protein-tyrosine kinases, through its SH2 domain, and acts as an adapter, mediating the association of the p110 catalytic unit to the plasma membrane. Indirectly regulates autophagy. Promotes nuclear translocation of XBP1 isoform 2 in a ER stress- and/or insulin-dependent manner during metabolic overloading in the liver and hence plays a role in glucose tolerance improvement.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.15E-01 Fold-change: 4.02E-03 Z-score: 4.09E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT/MRP1 signaling pathway | Activation | hsa04151 | |
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| SGC-7901/DDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| BGC-823/DDP cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometric cell cycle assay; Annexin V-FITC Apoptosis assay | |||
| Mechanism Description | HOTAIR was shown to directly bind to and inhibit miR126 expression and then to promote VEGFA and PIk3R2 expression and activate the PI3k/AkT/MRP1 pathway. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial adenocarcinoma [ICD-11: 2C76.0] | [2] | |||
| Sensitive Disease | Endometrial adenocarcinoma [ICD-11: 2C76.0] | |||
| Sensitive Drug | MTOR inhibitors | |||
| Molecule Alteration | Missense mutation | p.N561D (c.1681A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Whole-gene resequencing assay | |||
| Mechanism Description | The missense mutation p.N561D (c.1681A>G) in gene PIK3R2 cause the sensitivity of MTOR inhibitors by unusual activation of pro-survival pathway | |||
| Disease Class: Endometrial adenocarcinoma [ICD-11: 2C76.0] | [2] | |||
| Sensitive Disease | Endometrial adenocarcinoma [ICD-11: 2C76.0] | |||
| Sensitive Drug | MTOR inhibitors | |||
| Molecule Alteration | Missense mutation | p.A171V (c.512C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.15E-01; Fold-change: -1.82E-02; Z-score: -2.78E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.78E-02; Fold-change: 2.27E-01; Z-score: 7.67E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

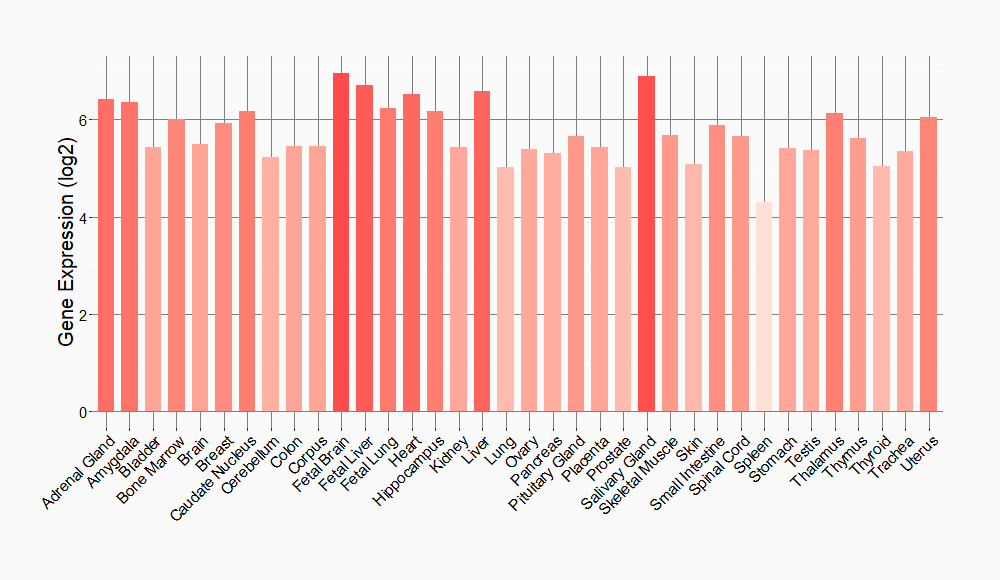
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
