Molecule Information
General Information of the Molecule (ID: Mol00501)
| Name |
Mitogen-activated protein kinase 3 (MAPK3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
MAP kinase 3; MAPK 3; ERT2; Extracellular signal-regulated kinase 1; ERK-1; Insulin-stimulated MAP2 kinase; MAP kinase isoform p44; p44-MAPK; Microtubule-associated protein 2 kinase; p44-ERK1; ERK1; PRKM3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
MAPK3
|
||||
| Gene ID | |||||
| Location |
chr16:30114105-30123506[-]
|
||||
| Sequence |
MAAAAAQGGGGGEPRRTEGVGPGVPGEVEMVKGQPFDVGPRYTQLQYIGEGAYGMVSSAY
DHVRKTRVAIKKISPFEHQTYCQRTLREIQILLRFRHENVIGIRDILRASTLEAMRDVYI VQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLINTTCDL KICDFGLARIADPEHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLS NRPIFPGKHYLDQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSD SKALDLLDRMLTFNPNKRITVEEALAHPYLEQYYDPTDEPVAEEPFTFAMELDDLPKERL KELIFQETARFQPGVLEAP Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, FRS2 or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3 or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6 or DUSP16) are other substrates which enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| MAPK2 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; EdU assay; Flow cytometry assay | |||
| Mechanism Description | BGC823/DDP and SGC7901/DDP cell presented lower miR-206 than parental cells, plus higher MAPk3 mRNA or protein. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Papillary thyroid carcinoma [ICD-11: 2D10.1] | [2] | |||
| Resistant Disease | Papillary thyroid carcinoma [ICD-11: 2D10.1] | |||
| Resistant Drug | Vemurafenib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ERK signaling pathway | Activation | hsa04210 | |
| mTOR signaling pathway | Activation | hsa04150 | ||
| In Vitro Model | BCPAP cells | Thyroid | Homo sapiens (Human) | CVCL_0153 |
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
Alamar blue assay | |||
| Mechanism Description | Resistance to vemurafenib in BCPAP appeared to be mediated by constitutive overexpression of phospho-ERK and by resistance to inhibition of both phospho-mTOR and phospho-S6 ribosomal protein after vemurafenib treatment. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.80E-01; Fold-change: -1.30E-01; Z-score: -3.50E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.23E-01; Fold-change: -9.58E-02; Z-score: -1.99E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
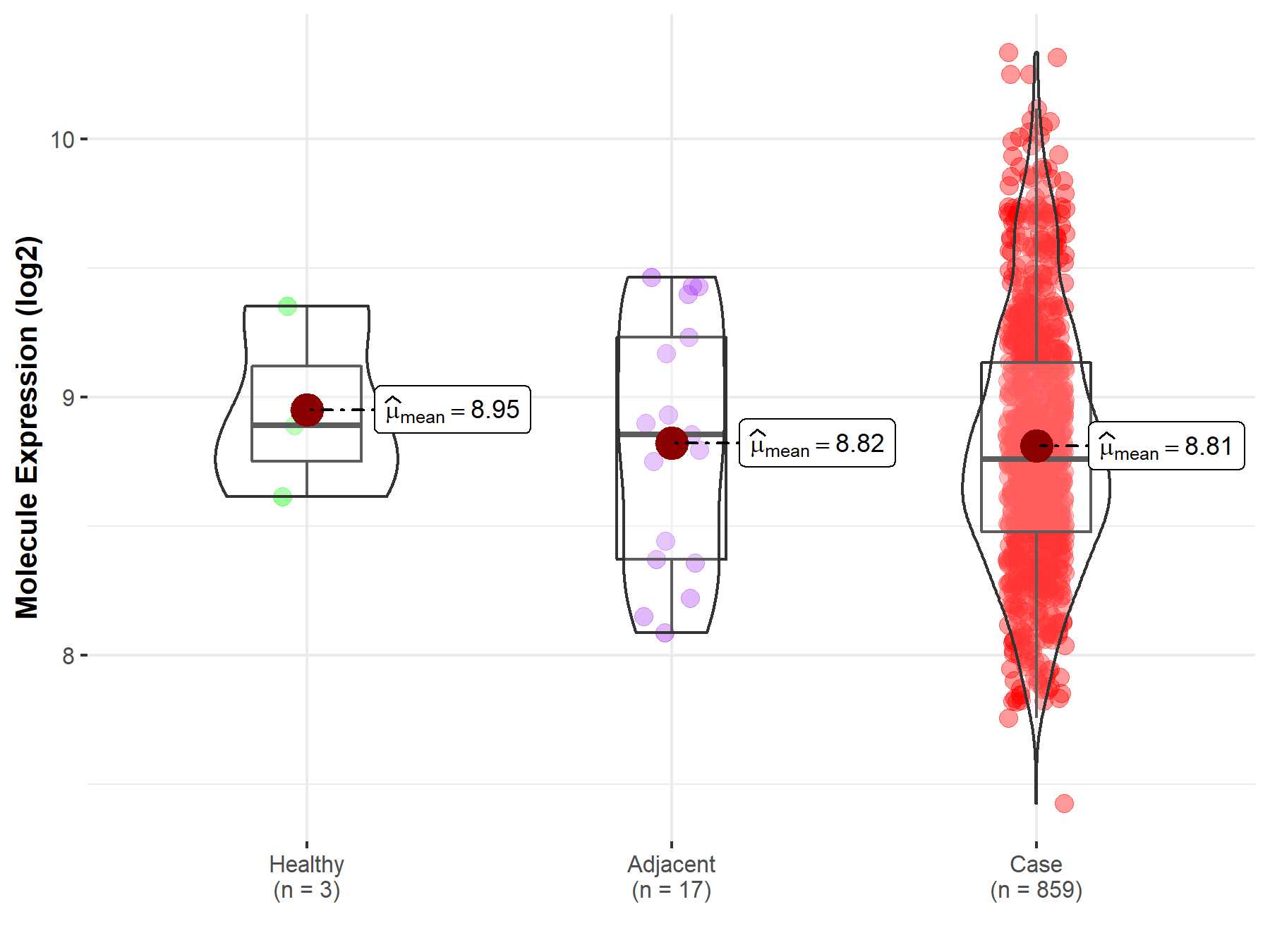
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.25E-03; Fold-change: -1.01E-01; Z-score: -4.34E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.47E-01; Fold-change: -2.50E-02; Z-score: -7.35E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
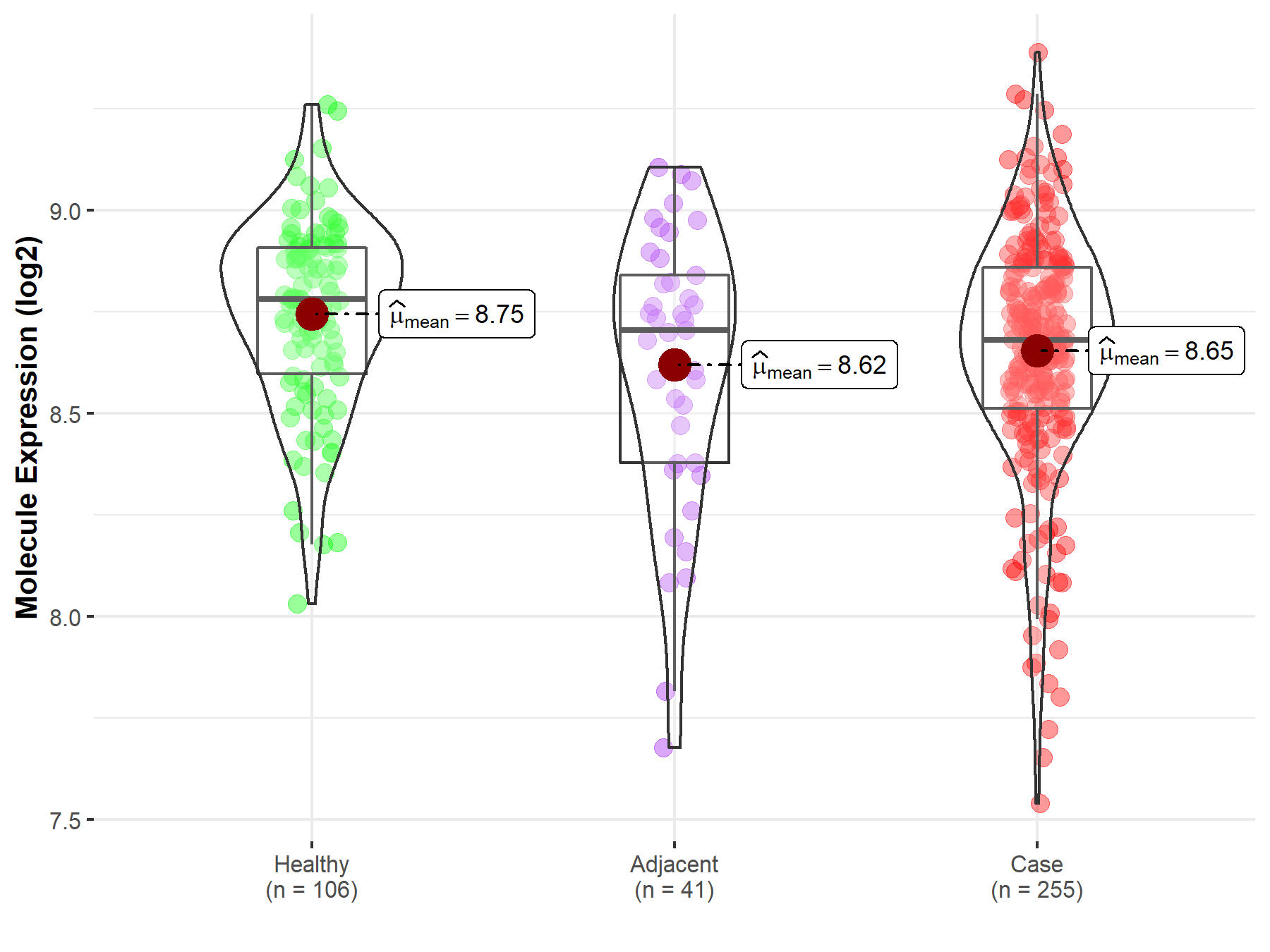
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

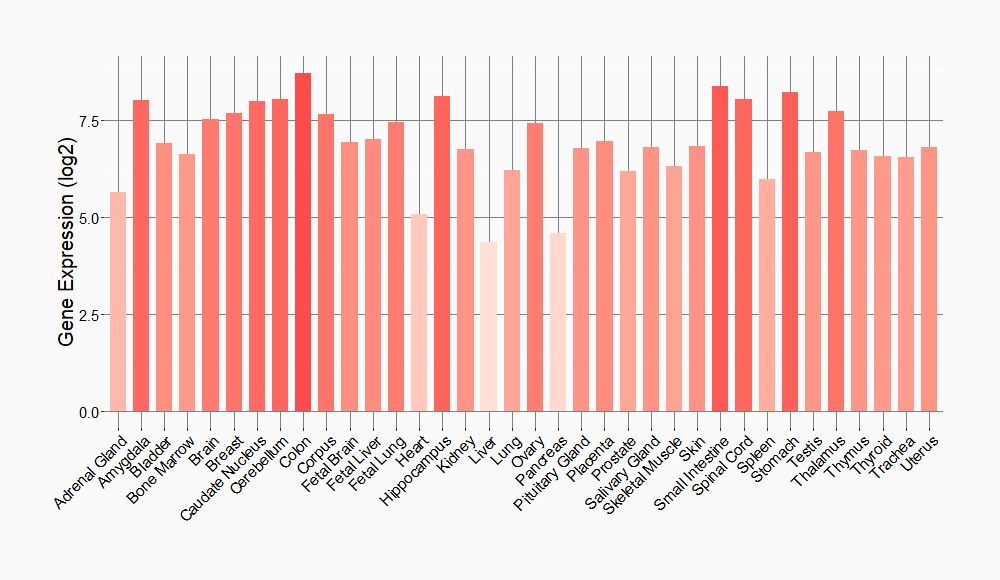
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
