Molecule Information
General Information of the Molecule (ID: Mol00456)
| Name |
Kelch-like ECH-associated protein 1 (KEAP1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Cytosolic inhibitor of Nrf2; INrf2; Kelch-like protein 19; INRF2; KIAA0132; KLHL19
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
KEAP1
|
||||
| Gene ID | |||||
| Location |
chr19:10486125-10503558[-]
|
||||
| Sequence |
MQPDPRPSGAGACCRFLPLQSQCPEGAGDAVMYASTECKAEVTPSQHGNRTFSYTLEDHT
KQAFGIMNELRLSQQLCDVTLQVKYQDAPAAQFMAHKVVLASSSPVFKAMFTNGLREQGM EVVSIEGIHPKVMERLIEFAYTASISMGEKCVLHVMNGAVMYQIDSVVRACSDFLVQQLD PSNAIGIANFAEQIGCVELHQRAREYIYMHFGEVAKQEEFFNLSHCQLVTLISRDDLNVR CESEVFHACINWVKYDCEQRRFYVQALLRAVRCHSLTPNFLQMQLQKCEILQSDSRCKDY LVKIFEELTLHKPTQVMPCRAPKVGRLIYTAGGYFRQSLSYLEAYNPSDGTWLRLADLQV PRSGLAGCVVGGLLYAVGGRNNSPDGNTDSSALDCYNPMTNQWSPCAPMSVPRNRIGVGV IDGHIYAVGGSHGCIHHNSVERYEPERDEWHLVAPMLTRRIGVGVAVLNRLLYAVGGFDG TNRLNSAECYYPERNEWRMITAMNTIRSGAGVCVLHNCIYAAGGYDGQDQLNSVERYDVE TETWTFVAPMKHRRSALGITVHQGRIYVLGGYDGHTFLDSVECYDPDTDTWSEVTRMTSG RSGVGVAVTMEPCRKQIDQQNCTC Click to Show/Hide
|
||||
| Function |
Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin ligase complex that regulates the response to oxidative stress by targeting NFE2L2/NRF2 for ubiquitination. KEAP1 acts as a key sensor of oxidative and electrophilic stress: in normal conditions, the BCR(KEAP1) complex mediates ubiquitination and degradation of NFE2L2/NRF2, a transcription factor regulating expression of many cytoprotective genes. In response to oxidative stress, different electrophile metabolites trigger non-enzymatic covalent modifications of highly reactive cysteine residues in KEAP1, leading to inactivate the ubiquitin ligase activity of the BCR(KEAP1) complex, promoting NFE2L2/NRF2 nuclear accumulation and expression of phase II detoxifying enzymes. In response to selective autophagy, KEAP1 is sequestered in inclusion bodies following its interaction with SQSTM1/p62, leading to inactivation of the BCR(KEAP1) complex and activation of NFE2L2/NRF2. The BCR(KEAP1) complex also mediates ubiquitination of SQSTM1/p62, increasing SQSTM1/p62 sequestering activity and degradation. The BCR(KEAP1) complex also targets BPTF and PGAM5 for ubiquitination and degradation by the proteasome.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.51E-03 Fold-change: 3.81E-02 Z-score: 3.34E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| Nrf2 signaling pathway | Inhibition | hsa05208 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Cells with kRAL overexpression exhibited a reversal in the resistance against 5-FU, with a significant decrease in the IC50 and a dramatic increase in cellular apoptosis, while silencing keap1 or ectopically expressing miR-141 partially rescued this effect. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [2] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.87E-02 Fold-change: 1.38E-01 Z-score: 3.12E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Knockdown miR-141 expression in 7901/DDP and 7901 cells could significantly improve cisplatin sensitivity. Over-expression of miR-141 resulted in (+) resistance to cisplatin in both gastric cancer cells. We also demonstrated that miR-141 directly targets kEAP1 by luciferase reporter assay, and that down-regulation of kEAP1 induces cisplatin resistance. Conversely, over-expression of kEAP1 significantly (+) cisplatin sensitivity. Our 75 pairs of tissues also showed that kEAP1 was significantly up-regulated in H. pylori-positive tissues. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [3] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| NF-kappaB signaling pathway | Activation | hsa04064 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| A2780 DDP cells | Ovary | Homo sapiens (Human) | CVCL_D619 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-141 regulates the expression of kEAP1 and that the repression of kEAP1 contributes to cisplatin resistance. Inhibition of NF-kB signaling enhances miR-141-mediated cisplatin sensitivity. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.87E-02; Fold-change: 6.21E-01; Z-score: 1.42E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.50E-04; Fold-change: 3.47E-01; Z-score: 1.20E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
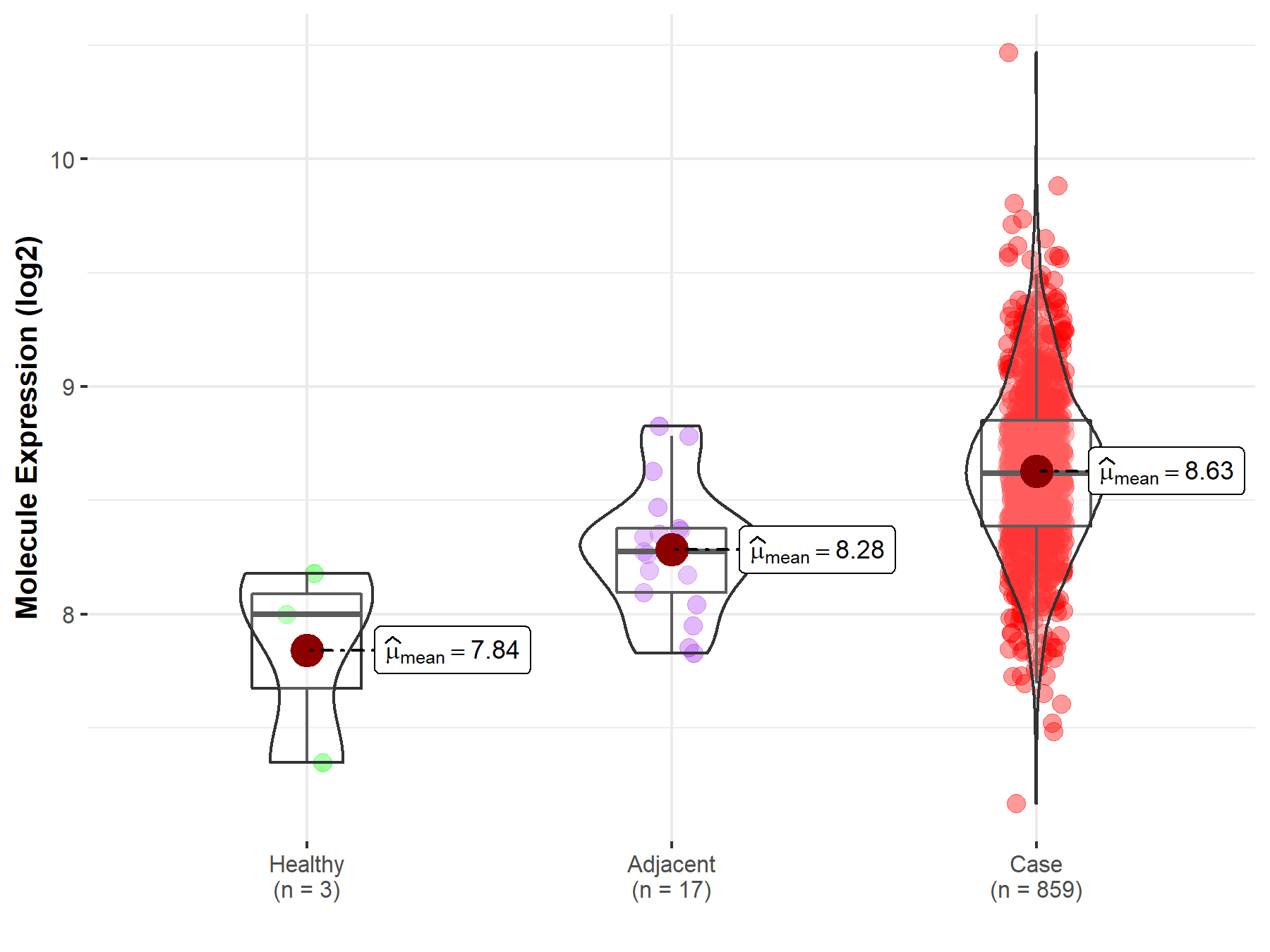
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.51E-03; Fold-change: 2.44E-01; Z-score: 6.18E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.06E-18; Fold-change: 3.16E-01; Z-score: 9.55E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 3.05E-01; Fold-change: 1.93E-01; Z-score: 7.66E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.87E-03; Fold-change: 6.30E-01; Z-score: 1.48E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.59E-04; Fold-change: 3.05E-01; Z-score: 1.00E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
