Molecule Information
General Information of the Molecule (ID: Mol00405)
| Name |
Histone deacetylase 3 (HDAC3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
HD3; Protein deacetylase HDAC3; Protein deacylase HDAC3; RPD3-2; SMAP45
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
HDAC3
|
||||
| Gene ID | |||||
| Location |
chr5:141620876-141636849[-]
|
||||
| Sequence |
MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN DKESDVEI Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Histone deacetylase that catalyzes the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4), and some other non-histone substrates. Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Participates in the BCL6 transcriptional repressor activity by deacetylating the H3 'Lys-27' (H3K27) on enhancer elements, antagonizing EP300 acetyltransferase activity and repressing proximal gene expression. Acts as a molecular chaperone for shuttling phosphorylated NR2C1 to PML bodies for sumoylation. Contributes, together with XBP1 isoform 1, to the activation of NFE2L2-mediated HMOX1 transcription factor gene expression in a PI(3)K/mTORC2/Akt-dependent signaling pathway leading to endothelial cell (EC) survival under disturbed flow/oxidative stress. Regulates both the transcriptional activation and repression phases of the circadian clock in a deacetylase activity-independent manner. During the activation phase, promotes the accumulation of ubiquitinated ARNTL/BMAL1 at the E-boxes and during the repression phase, blocks FBXL3-mediated CRY1/2 ubiquitination and promotes the interaction of CRY1 and ARNTL/BMAL1. The NCOR1-HDAC3 complex regulates the circadian expression of the core clock gene ARTNL/BMAL1 and the genes involved in lipid metabolism in the liver. Also functions as deacetylase for non-histone targets, such as KAT5, MEF2D, MAPK14 and RARA. Serves as a corepressor of RARA, mediating its deacetylation and repression, leading to inhibition of RARE DNA element binding. In association with RARA, plays a role in the repression of microRNA-10a and thereby in the inflammatory response. In addition to protein deacetylase activity, also acts as protein-lysine deacylase by recognizing other acyl groups: catalyzes removal of (2E)-butenoyl (crotonyl) and 2-hydroxyisobutanoyl (2-hydroxyisobutyryl) acyl groups from lysine residues, leading to protein decrotonylation and de-2-hydroxyisobutyrylation, respectively. Catalyzes decrotonylation of MAPRE1/EB1.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [1] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Melanoma [ICD-11: 2C30] | |||
| The Specified Disease | Melanoma | |||
| The Studied Tissue | Skin | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.19E-01 Fold-change: -1.04E-02 Z-score: -8.20E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 |
| Malme3M cells | Skin | Homo sapiens (Human) | CVCL_1438 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-326, which forms a negative feedback regulatory loop with HDAC3, regulates the invasion and the metastatic potential of cancer cells and tumor-induced angiogenesis in response to anti-cancer drugs. miR-200b, miR-217, and miR-335, which form a positive feedback loop with HDAC3, confer sensitivity to anti-cancer drugs. We show that CAGE, reported to form a feedback loop with miR-200b, serves as a downstream target of HDAC3 and miR-326. In this study, we show that the regulation of the miR-326/HDAC3 axis can be employed for the development of anti-cancer therapeutics. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.19E-01; Fold-change: -2.36E-02; Z-score: -6.08E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

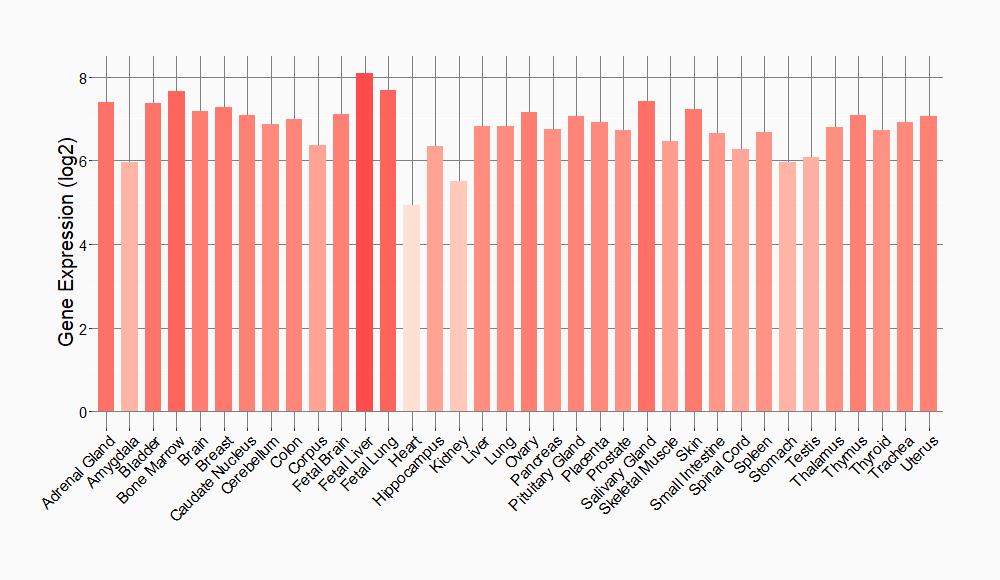
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
