Molecule Information
General Information of the Molecule (ID: Mol00361)
| Name |
Histone-lysine N-methyltransferase EZH1 (EZH1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
ENX-2; Enhancer of zeste homolog 1; KIAA0388
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
EZH1
|
||||
| Gene ID | |||||
| Location |
chr17:42700275-42745049[-]
|
||||
| Sequence |
MEIPNPPTSKCITYWKRKVKSEYMRLRQLKRLQANMGAKALYVANFAKVQEKTQILNEEW
KKLRVQPVQSMKPVSGHPFLKKCTIESIFPGFASQHMLMRSLNTVALVPIMYSWSPLQQN FMVEDETVLCNIPYMGDEVKEEDETFIEELINNYDGKVHGEEEMIPGSVLISDAVFLELV DALNQYSDEEEEGHNDTSDGKQDDSKEDLPVTRKRKRHAIEGNKKSSKKQFPNDMIFSAI ASMFPENGVPDDMKERYRELTEMSDPNALPPQCTPNIDGPNAKSVQREQSLHSFHTLFCR RCFKYDCFLHPFHATPNVYKRKNKEIKIEPEPCGTDCFLLLEGAKEYAMLHNPRSKCSGR RRRRHHIVSASCSNASASAVAETKEGDSDRDTGNDWASSSSEANSRCQTPTKQKASPAPP QLCVVEAPSEPVEWTGAEESLFRVFHGTYFNNFCSIARLLGTKTCKQVFQFAVKESLILK LPTDELMNPSQKKKRKHRLWAAHCRKIQLKKDNSSTQVYNYQPCDHPDRPCDSTCPCIMT QNFCEKFCQCNPDCQNRFPGCRCKTQCNTKQCPCYLAVRECDPDLCLTCGASEHWDCKVV SCKNCSIQRGLKKHLLLAPSDVAGWGTFIKESVQKNEFISEYCGELISQDEADRRGKVYD KYMSSFLFNLNNDFVVDATRKGNKIRFANHSVNPNCYAKVVMVNGDHRIGIFAKRAIQAG EELFFDYRYSQADALKYVGIERETDVL Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Polycomb group (PcG) protein. Catalytic subunit of the PRC2/EED-EZH1 complex, which methylates 'Lys-27' of histone H3, leading to transcriptional repression of the affected target gene. Able to mono-, di- and trimethylate 'Lys-27' of histone H3 to form H3K27me1, H3K27me2 and H3K27me3, respectively. Required for embryonic stem cell derivation and self-renewal, suggesting that it is involved in safeguarding embryonic stem cell identity. Compared to EZH2-containing complexes, it is less abundant in embryonic stem cells, has weak methyltransferase activity and plays a less critical role in forming H3K27me3, which is required for embryonic stem cell identity and proper differentiation.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [1] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Erlotinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.76E-01 Fold-change: 4.58E-03 Z-score: 5.60E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR17-5p down-regulation contributes to erlotinib resistance in non-small cell lung cancer cells, miR17-5p could inhibit the mRNA and protein levels of EZH1. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.76E-01; Fold-change: -4.50E-02; Z-score: -1.18E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.66E-01; Fold-change: -1.27E-02; Z-score: -3.58E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
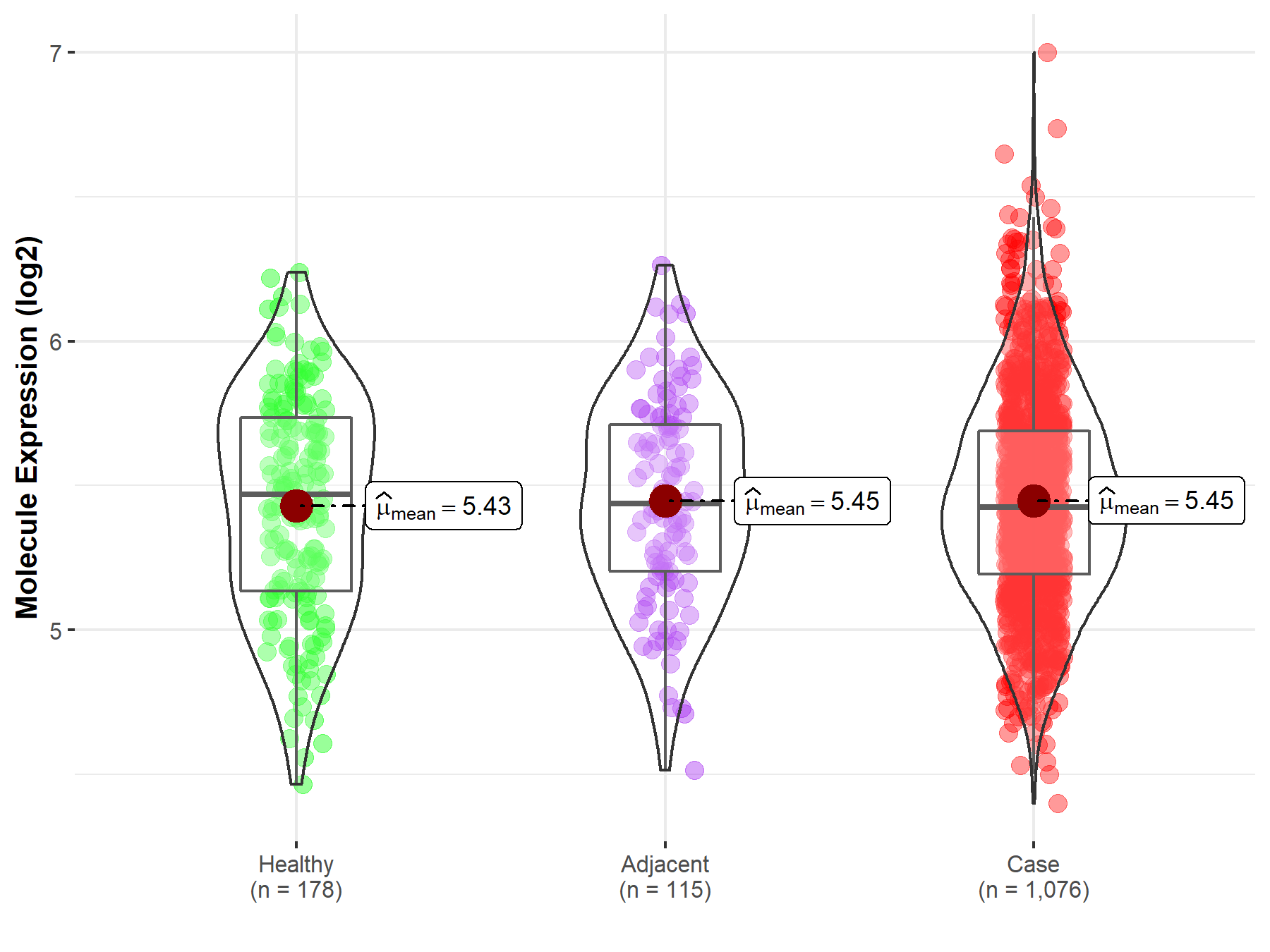
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

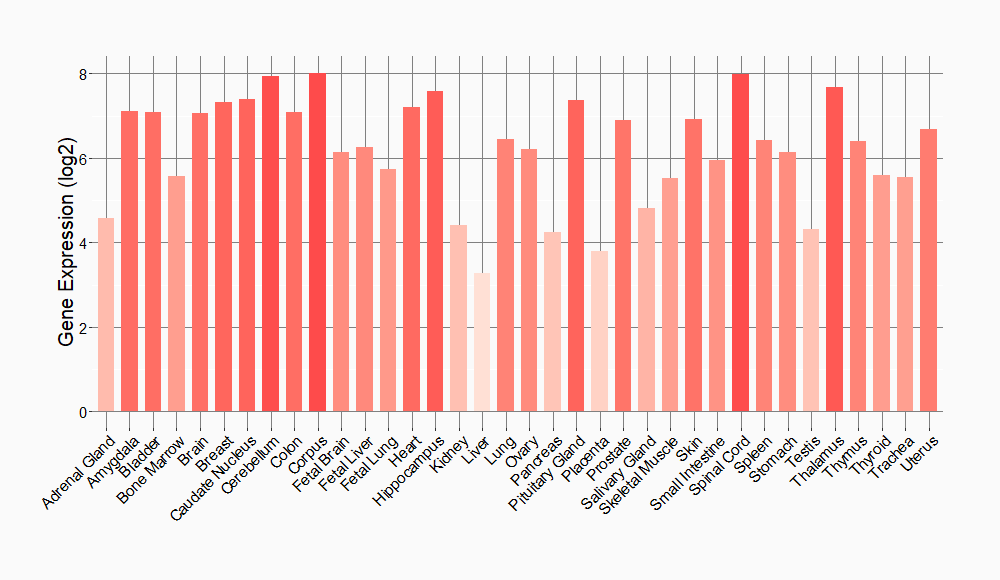
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
