Molecule Information
General Information of the Molecule (ID: Mol00330)
| Name |
DNA polymerase theta (POLQ)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
DNA polymerase eta; POLH
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
POLQ
|
||||
| Gene ID | |||||
| Location |
chr3:121431431-121545988[-]
|
||||
| Sequence |
MNLLRRSGKRRRSESGSDSFSGSGGDSSASPQFLSGSVLSPPPGLGRCLKAAAAGECKPT
VPDYERDKLLLANWGLPKAVLEKYHSFGVKKMFEWQAECLLLGQVLEGKNLVYSAPTSAG KTLVAELLILKRVLEMRKKALFILPFVSVAKEKKYYLQSLFQEVGIKVDGYMGSTSPSRH FSSLDIAVCTIERANGLINRLIEENKMDLLGMVVVDELHMLGDSHRGYLLELLLTKICYI TRKSASCQADLASSLSNAVQIVGMSATLPNLELVASWLNAELYHTDFRPVPLLESVKVGN SIYDSSMKLVREFEPMLQVKGDEDHVVSLCYETICDNHSVLLFCPSKKWCEKLADIIARE FYNLHHQAEGLVKPSECPPVILEQKELLEVMDQLRRLPSGLDSVLQKTVPWGVAFHHAGL TFEERDIIEGAFRQGLIRVLAATSTLSSGVNLPARRVIIRTPIFGGRPLDILTYKQMVGR AGRKGVDTVGESILICKNSEKSKGIALLQGSLKPVRSCLQRREGEEVTGSMIRAILEIIV GGVASTSQDMHTYAACTFLAASMKEGKQGIQRNQESVQLGAIEACVMWLLENEFIQSTEA SDGTEGKVYHPTHLGSATLSSSLSPADTLDIFADLQRAMKGFVLENDLHILYLVTPMFED WTTIDWYRFFCLWEKLPTSMKRVAELVGVEEGFLARCVKGKVVARTERQHRQMAIHKRFF TSLVLLDLISEVPLREINQKYGCNRGQIQSLQQSAAVYAGMITVFSNRLGWHNMELLLSQ FQKRLTFGIQRELCDLVRVSLLNAQRARVLYASGFHTVADLARANIVEVEVILKNAVPFK SARKAVDEEEEAVEERRNMRTIWVTGRKGLTEREAAALIVEEARMILQQDLVEMGVQWNP CALLHSSTCSLTHSESEVKEHTFISQTKSSYKKLTSKNKSNTIFSDSYIKHSPNIVQDLN KSREHTSSFNCNFQNGNQEHQTCSIFRARKRASLDINKEKPGASQNEGKTSDKKVVQTFS QKTKKAPLNFNSEKMSRSFRSWKRRKHLKRSRDSSPLKDSGACRIHLQGQTLSNPSLCED PFTLDEKKTEFRNSGPFAKNVSLSGKEKDNKTSFPLQIKQNCSWNITLTNDNFVEHIVTG SQSKNVTCQATSVVSEKGRGVAVEAEKINEVLIQNGSKNQNVYMKHHDIHPINQYLRKQS HEQTSTITKQKNIIERQMPCEAVSSYINRDSNVTINCERIKLNTEENKPSHFQALGDDIS RTVIPSEVLPSAGAFSKSEGQHENFLNISRLQEKTGTYTTNKTKNNHVSDLGLVLCDFED SFYLDTQSEKIIQQMATENAKLGAKDTNLAAGIMQKSLVQQNSMNSFQKECHIPFPAEQH PLGATKIDHLDLKTVGTMKQSSDSHGVDILTPESPIFHSPILLEENGLFLKKNEVSVTDS QLNSFLQGYQTQETVKPVILLIPQKRTPTGVEGECLPVPETSLNMSDSLLFDSFSDDYLV KEQLPDMQMKEPLPSEVTSNHFSDSLCLQEDLIKKSNVNENQDTHQQLTCSNDESIIFSE MDSVQMVEALDNVDIFPVQEKNHTVVSPRALELSDPVLDEHHQGDQDGGDQDERAEKSKL TGTRQNHSFIWSGASFDLSPGLQRILDKVSSPLENEKLKSMTINFSSLNRKNTELNEEQE VISNLETKQVQGISFSSNNEVKSKIEMLENNANHDETSSLLPRKESNIVDDNGLIPPTPI PTSASKLTFPGILETPVNPWKTNNVLQPGESYLFGSPSDIKNHDLSPGSRNGFKDNSPIS DTSFSLQLSQDGLQLTPASSSSESLSIIDVASDQNLFQTFIKEWRCKKRFSISLACEKIR SLTSSKTATIGSRFKQASSPQEIPIRDDGFPIKGCDDTLVVGLAVCWGGRDAYYFSLQKE QKHSEISASLVPPSLDPSLTLKDRMWYLQSCLRKESDKECSVVIYDFIQSYKILLLSCGI SLEQSYEDPKVACWLLDPDSQEPTLHSIVTSFLPHELPLLEGMETSQGIQSLGLNAGSEH SGRYRASVESILIFNSMNQLNSLLQKENLQDVFRKVEMPSQYCLALLELNGIGFSTAECE SQKHIMQAKLDAIETQAYQLAGHSFSFTSSDDIAEVLFLELKLPPNREMKNQGSKKTLGS TRRGIDNGRKLRLGRQFSTSKDVLNKLKALHPLPGLILEWRRITNAITKVVFPLQREKCL NPFLGMERIYPVSQSHTATGRITFTEPNIQNVPRDFEIKMPTLVGESPPSQAVGKGLLPM GRGKYKKGFSVNPRCQAQMEERAADRGMPFSISMRHAFVPFPGGSILAADYSQLELRILA HLSHDRRLIQVLNTGADVFRSIAAEWKMIEPESVGDDLRQQAKQICYGIIYGMGAKSLGE QMGIKENDAACYIDSFKSRYTGINQFMTETVKNCKRDGFVQTILGRRRYLPGIKDNNPYR KAHAERQAINTIVQGSAADIVKIATVNIQKQLETFHSTFKSHGHREGMLQSDQTGLSRKR KLQGMFCPIRGGFFILQLHDELLYEVAEEDVVQVAQIVKNEMESAVKLSVKLKVKVKIGA SWGELKDFDV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
DNA polymerase that promotes microhomology-mediated end-joining (MMEJ), an alternative non-homologous end-joining (NHEJ) machinery triggered in response to double-strand breaks in DNA. MMEJ is an error-prone repair pathway that produces deletions of sequences from the strand being repaired and promotes genomic rearrangements, such as telomere fusions, some of them leading to cellular transformation. POLQ acts as an inhibitor of homology-recombination repair (HR) pathway by limiting RAD51 accumulation at resected ends. POLQ-mediated MMEJ may be required to promote the survival of cells with a compromised HR repair pathway, thereby preventing genomic havoc by resolving unrepaired lesions. The polymerase acts by binding directly the 2 ends of resected double-strand breaks, allowing microhomologous sequences in the overhangs to form base pairs. It then extends each strand from the base-paired region using the opposing overhang as a template. Requires partially resected DNA containing 2 to 6 base pairs of microhomology to perform MMEJ. The polymerase activity is highly promiscuous: unlike most polymerases, promotes extension of ssDNA and partial ssDNA (pssDNA) substrates. Also exhibits low-fidelity DNA synthesis, translesion synthesis and lyase activity, and it is implicated in interstrand-cross-link repair, base excision repair and DNA end-joining. Involved in somatic hypermutation of immunoglobulin genes, a process that requires the activity of DNA polymerases to ultimately introduce mutations at both A/T and C/G base pairs.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [1] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| OV2008 cells | Ovary | Homo sapiens (Human) | CVCL_0473 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | There is an elevated expression of DNA polymerase Eta (Pol Eta) in ovarian CSCs isolated from both ovarian cancer cell lines and primary tumors, indicating that CSCs may have intrinsically (+) translesion DNA synthesis (TLS). Down-regulation of Pol Eta blocked cisplatin-induced CSC enrichment both in vitro and in vivo through the enhancement of cisplatin-induced apoptosis in CSCs, indicating that Pol Eta-mediated TLS contributes to the survival of CSCs upon cisplatin treatment. Furthermore, our data demonstrated a depletion of miR-93 in ovarian CSCs. Enforced expression of miR-93 in ovarian CSCs reduced Pol Eta expression and increased their sensitivity to cisplatin. Taken together, our data suggest that ovarian CSCs have intrinsically (+) Pol Eta-mediated TLS, allowing CSCs to survive cisplatin treatment, leading to tumor relapse. Targeting Pol Eta, probably through enhancement of miR-93 expression, might be exploited as a strategy to increase the efficacy of cisplatin treatment. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.78E-07; Fold-change: 1.33E+00; Z-score: 4.37E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.21E-05; Fold-change: 9.14E-01; Z-score: 2.21E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
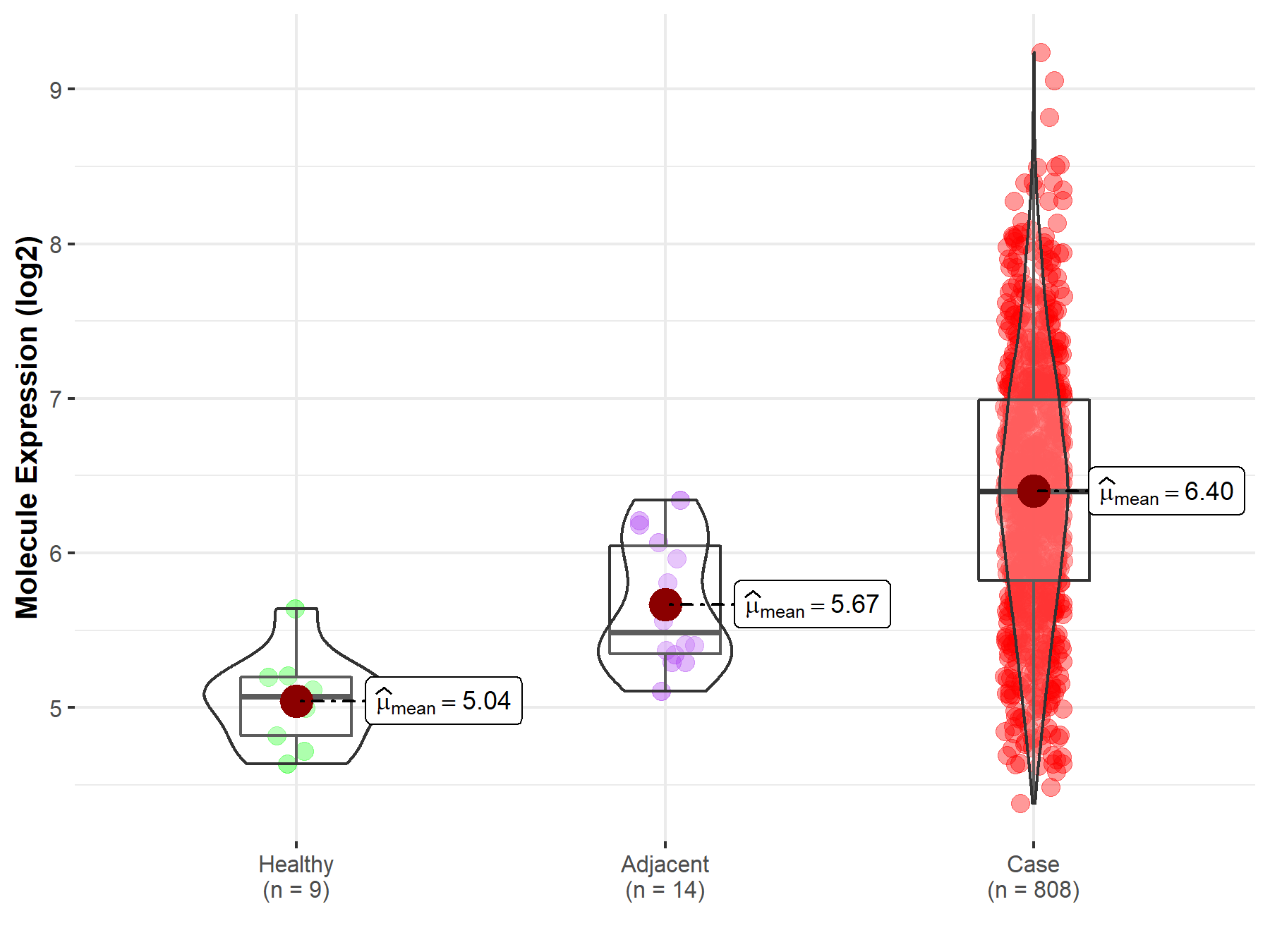
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

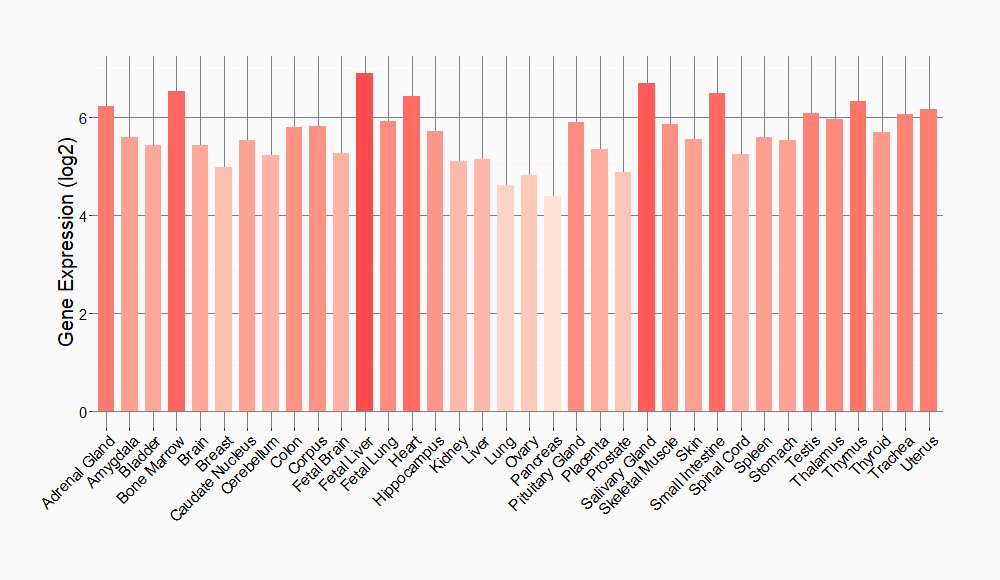
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
