Molecule Information
General Information of the Molecule (ID: Mol00320)
| Name |
m7GpppN-mRNA hydrolase (DCP2)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Nucleoside diphosphate-linked moiety X motif 20; Nudix motif 20; mRNA-decapping enzyme 2; hDpc; NUDT20
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
DCP2
|
||||
| Gene ID | |||||
| Location |
chr5:112976702-113022195[+]
|
||||
| Sequence |
METKRVEIPGSVLDDLCSRFILHIPSEERDNAIRVCFQIELAHWFYLDFYMQNTPGLPQC
GIRDFAKAVFSHCPFLLPQGEDVEKVLDEWKEYKMGVPTYGAIILDETLENVLLVQGYLA KSGWGFPKGKVNKEEAPHDCAAREVFEETGFDIKDYICKDDYIELRINDQLARLYIIPGI PKDTKFNPKTRREIRNIEWFSIEKLPCHRNDMTPKSKLGLAPNKFFMAIPFIRPLRDWLS RRFGDSSDSDNGFSSTGSTPAKPTVEKLSRTKFRHSQQLFPDGSPGDQWVKHRQPLQQKP YNNHSEMSDLLKGKNQSMRGNGRKQYQDSPNQKKRTNGLQPAKQQNSLMKCEKKLHPRKL QDNFETDAVYDLPSSSEDQLLEHAEGQPVACNGHCKFPFSSRAFLSFKFDHNAIMKILDL Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Decapping metalloenzyme that catalyzes the cleavage of the cap structure on mRNAs. Removes the 7-methyl guanine cap structure from mRNA molecules, yielding a 5'-phosphorylated mRNA fragment and 7m-GDP. Necessary for the degradation of mRNAs, both in normal mRNA turnover and in nonsense-mediated mRNA decay. Plays a role in replication-dependent histone mRNA degradation. Has higher activity towards mRNAs that lack a poly(A) tail. Has no activity towards a cap structure lacking an RNA moiety. The presence of a N(6)-methyladenosine methylation at the second transcribed position of mRNAs (N(6),2'-O-dimethyladenosine cap; m6A(m)) provides resistance to DCP2-mediated decapping. Blocks autophagy in nutrient-rich conditions by repressing the expression of ATG-related genes through degradation of their transcripts.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [1] | |||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Resistant Drug | Methotrexate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-06 Fold-change: 3.03E-02 Z-score: 4.81E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | The endogenous overexpression of CDS2, DCP2, HSPC159, MYST3 and SLC4A4 in MTX-resistant HT29 cells. Inhibition of miR-224 with anti-miR224 produced an increase in the mRNA levels of CDS2, HSPC159, MYST3 and SLC4A4. Decreased mRNA levels of SLC4A4, CDS2 and HSPC159 cause an increase in MTX sensitivity in HT29 cells. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-06; Fold-change: 4.89E-02; Z-score: 2.02E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.68E-03; Fold-change: -1.31E-01; Z-score: -3.47E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
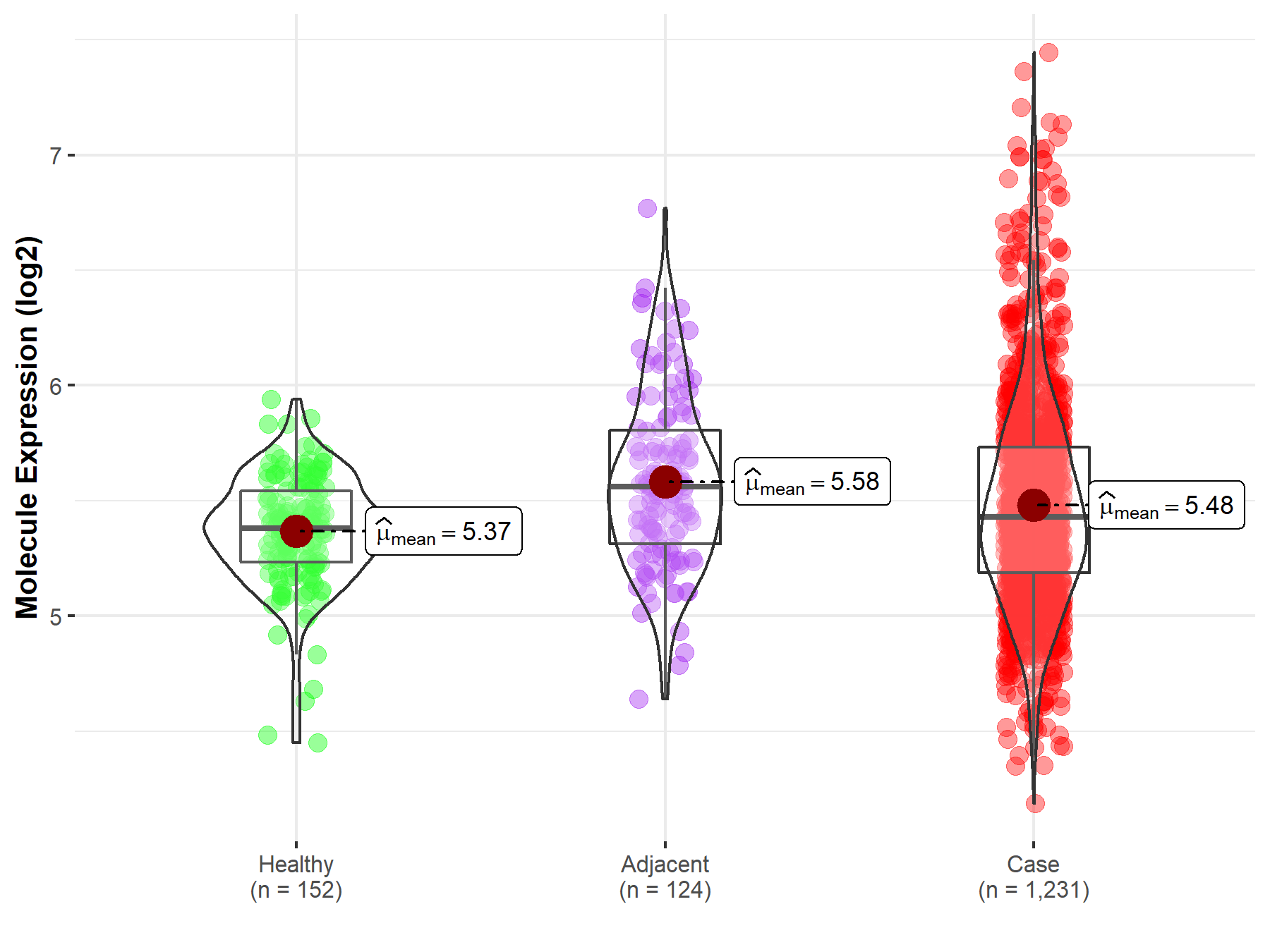
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
