Molecule Information
General Information of the Molecule (ID: Mol00292)
| Name |
Matrix protein P1 (HSPD1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
60 kDa chaperonin; Chaperonin 60; CPN60; Heat shock protein 60; HSP-60; Hsp60; HuCHA60; Mitochondrial matrix protein P1; P60 lymphocyte protein; HSP60
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
HSPD1
|
||||
| Gene ID | |||||
| Location |
chr2:197486584-197516737[-]
|
||||
| Sequence |
MLRLPTVFRQMRPVSRVLAPHLTRAYAKDVKFGADARALMLQGVDLLADAVAVTMGPKGR
TVIIEQSWGSPKVTKDGVTVAKSIDLKDKYKNIGAKLVQDVANNTNEEAGDGTTTATVLA RSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELKKQSKPVTTPEEIAQVATISANGDK EIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEGMKFDRGYISPYFINTSKGQKCEFQ DAYVLLSEKKISSIQSIVPALEIANAHRKPLVIIAEDVDGEALSTLVLNRLKVGLQVVAV KAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLEDVQPHDLGKVGEVIVTKDDAMLLKG KGDKAQIEKRIQEIIEQLDVTTSEYEKEKLNERLAKLSDGVAVLKVGGTSDVEVNEKKDR VTDALNATRAAVEEGIVLGGGCALLRCIPALDSLTPANEDQKIGIEIIKRTLKIPAMTIA KNAGVEGSLIVEKIMQSSSEVGYDAMAGDFVNMVEKGIIDPTKVVRTALLDAAGVASLLT TAEVVVTEIPKEEKDPGMGAMGGMGGGMGGGMF Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Chaperonin implicated in mitochondrial protein import and macromolecular assembly. Together with Hsp10, facilitates the correct folding of imported proteins. May also prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions in the mitochondrial matrix. The functional units of these chaperonins consist of heptameric rings of the large subunit Hsp60, which function as a back-to-back double ring. In a cyclic reaction, Hsp60 ring complexes bind one unfolded substrate protein per ring, followed by the binding of ATP and association with 2 heptameric rings of the co-chaperonin Hsp10. This leads to sequestration of the substrate protein in the inner cavity of Hsp60 where, for a certain period of time, it can fold undisturbed by other cell components. Synchronous hydrolysis of ATP in all Hsp60 subunits results in the dissociation of the chaperonin rings and the release of ADP and the folded substrate protein (Probable).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [1] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.17E-02 Fold-change: -3.08E-01 Z-score: -2.84E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | COC1 cells | Ovary | Homo sapiens (Human) | CVCL_6891 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulations of PkM2 and HSPD1 involved in MDR in ovarian cancer. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.17E-02; Fold-change: -2.02E+00; Z-score: -1.36E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.34E-03; Fold-change: 3.38E-01; Z-score: 5.76E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
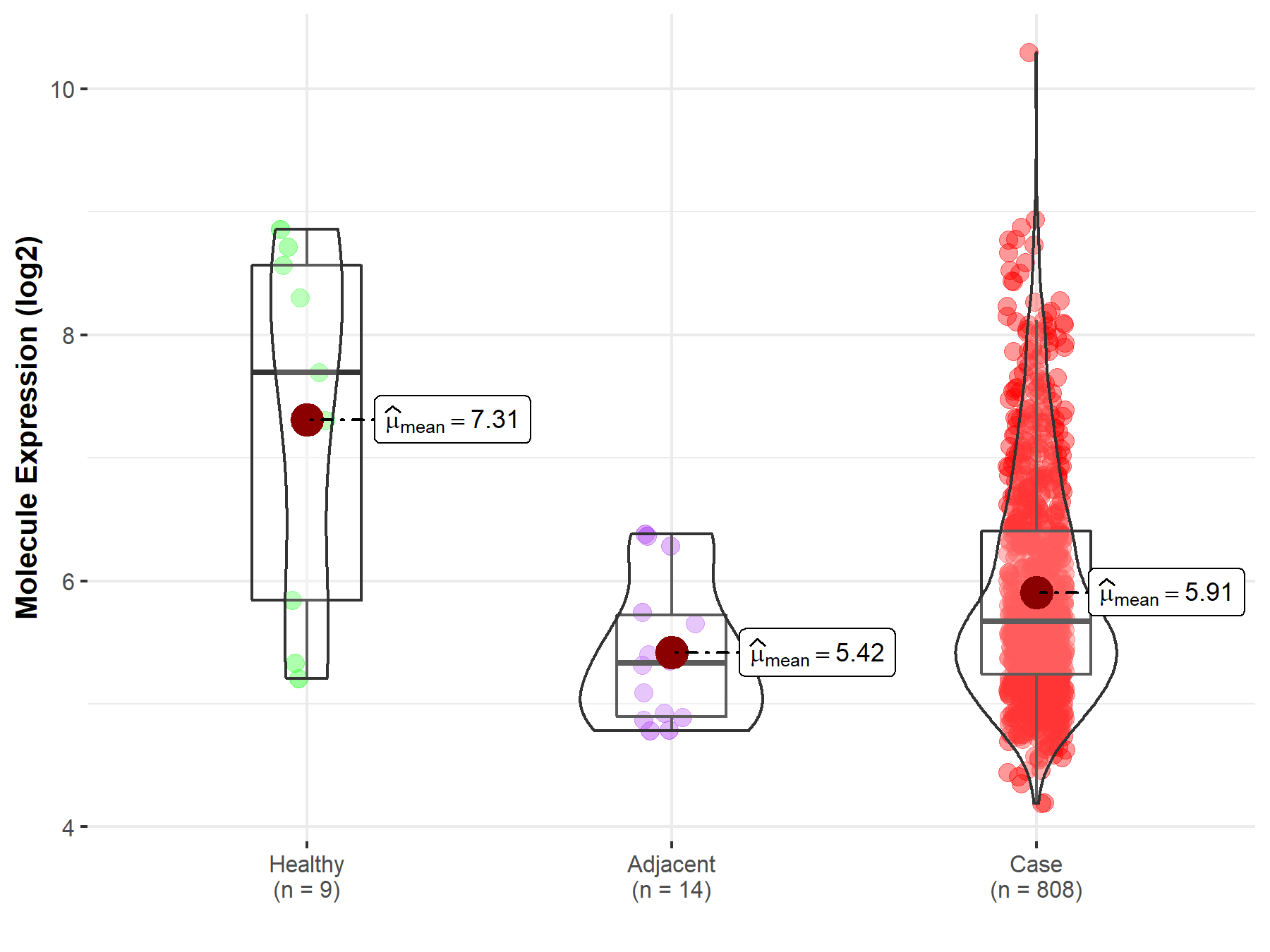
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

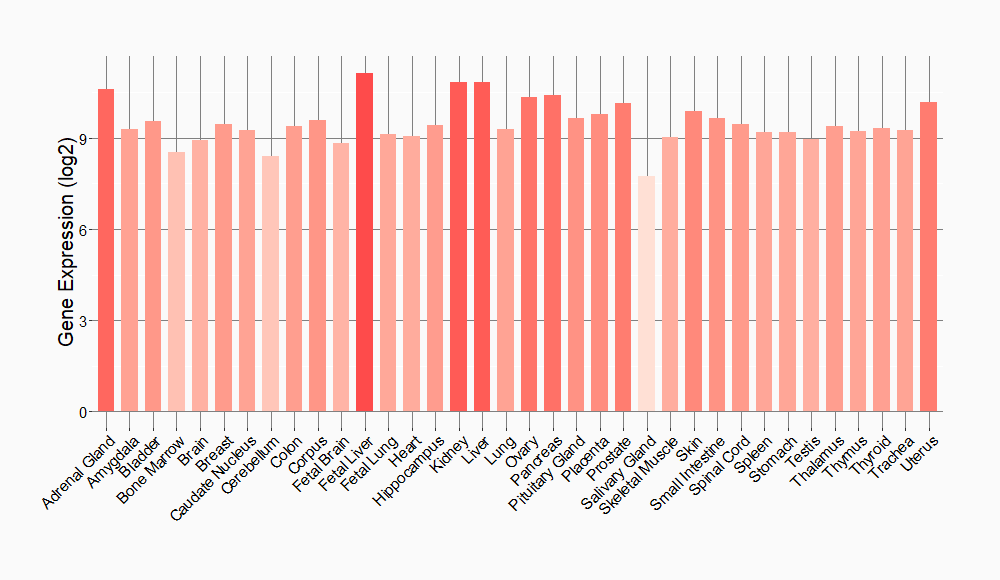
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
