Molecule Information
General Information of the Molecule (ID: Mol00249)
| Name |
Beclin 1-associated autophagy-related key regulator (ATG14)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Barkor; Autophagy-related protein 14-like protein; Atg14L; ATG14L; KIAA0831
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATG14
|
||||
| Gene ID | |||||
| Location |
chr14:55366391-55411830[-]
|
||||
| Sequence |
MASPSGKGARALEAPGCGPRPLARDLVDSVDDAEGLYVAVERCPLCNTTRRRLTCAKCVQ
SGDFVYFDGRDRERFIDKKERLSRLKSKQEEFQKEVLKAMEGKWITDQLRWKIMSCKMRI EQLKQTICKGNEEMEKNSEGLLKTKEKNQKLYSRAQRHQEKKEKIQRHNRKLGDLVEKKT IDLRSHYERLANLRRSHILELTSVIFPIEEVKTGVRDPADVSSESDSAMTSSTVSKLAEA RRTTYLSGRWVCDDHNGDTSISITGPWISLPNNGDYSAYYSWVEEKKTTQGPDMEQSNPA YTISAALCYATQLVNILSHILDVNLPKKLCNSEFCGENLSKQKFTRAVKKLNANILYLCF SQHVNLDQLQPLHTLRNLMYLVSPSSEHLGRSGPFEVRADLEESMEFVDPGVAGESDESG DERVSDEETDLGTDWENLPSPRFCDIPSQSVEVSQSQSTQASPPIASSSAGGMISSAAAS VTSWFKAYTGHR Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Required for both basal and inducible autophagy. Determines the localization of the autophagy-specific PI3-kinase complex PI3KC3-C1. Plays a role in autophagosome formation and MAP1LC3/LC3 conjugation to phosphatidylethanolamine. Promotes BECN1 translocation from the trans-Golgi network to autophagosomes. Enhances PIK3C3 activity in a BECN1-dependent manner. Essential for the autophagy-dependent phosphorylation of BECN1. Stimulates the phosphorylation of BECN1, but suppresses the phosphorylation PIK3C3 by AMPK. Binds to STX17-SNAP29 binary t-SNARE complex on autophagosomes and primes it for VAMP8 interaction to promote autophagosome-endolysosome fusion. Modulates the hepatic lipid metabolism.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [1] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Melphalan | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Multiple myeloma [ICD-11: 2A83] | |||
| The Specified Disease | Myeloma | |||
| The Studied Tissue | Bone marrow | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.37E-01 Fold-change: 1.75E-02 Z-score: 8.21E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell autophagy | Activation | hsa04140 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 |
| LP1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0012 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Linc00515 enhanced autophagy and chemoresistance of melphalan-resistant myeloma by directly inhibiting miR-140-5p, which elevated ATG14 level. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [1] | |||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Sensitive Drug | Melphalan | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell autophagy | Activation | hsa04140 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 |
| LP1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0012 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Linc00515 enhanced autophagy and chemoresistance of melphalan-resistant myeloma by directly inhibiting miR-140-5p, which elevated ATG14 level. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.37E-01; Fold-change: 7.45E-02; Z-score: 2.46E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
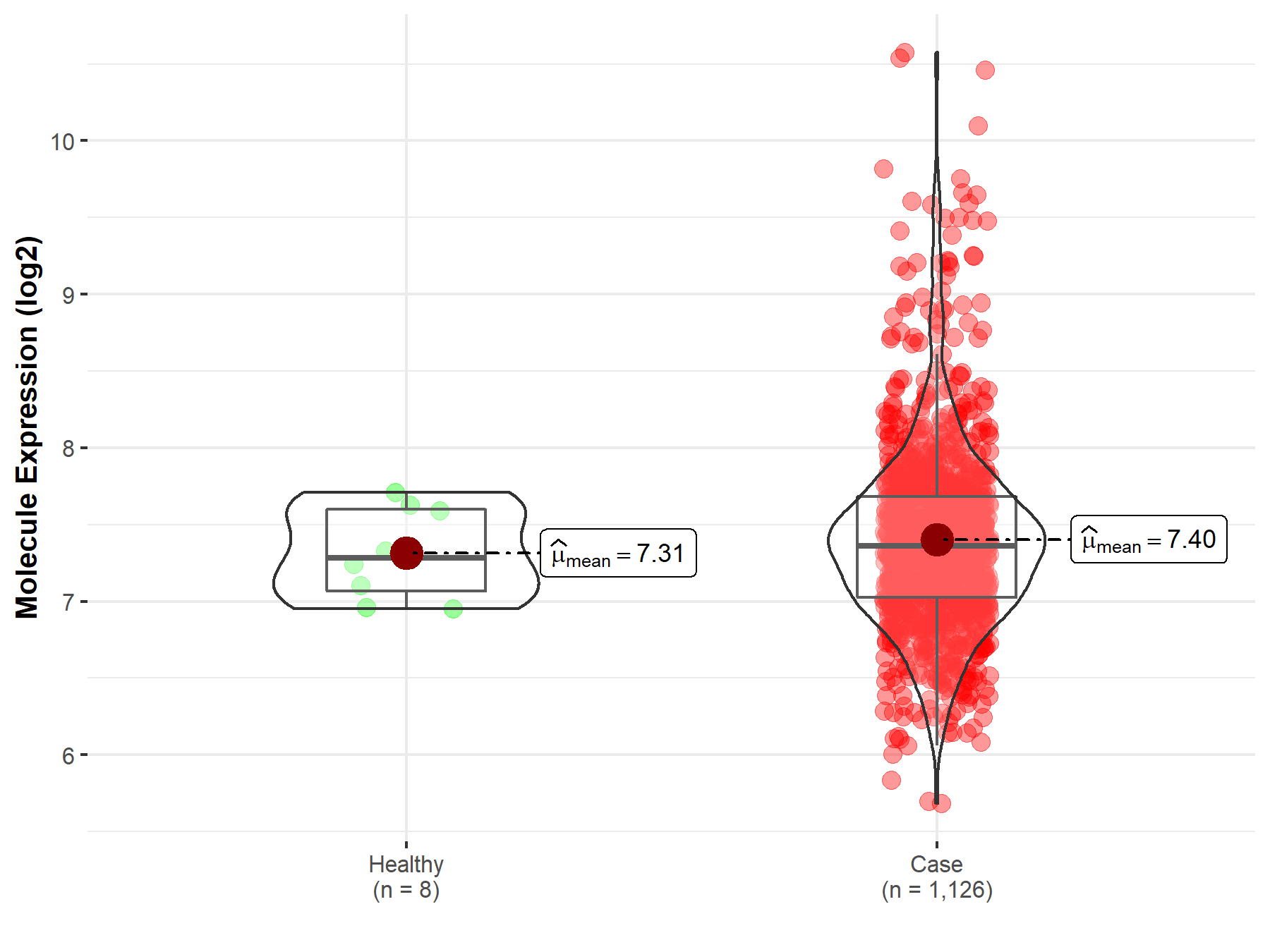
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.58E-01; Fold-change: 4.58E-01; Z-score: 5.23E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
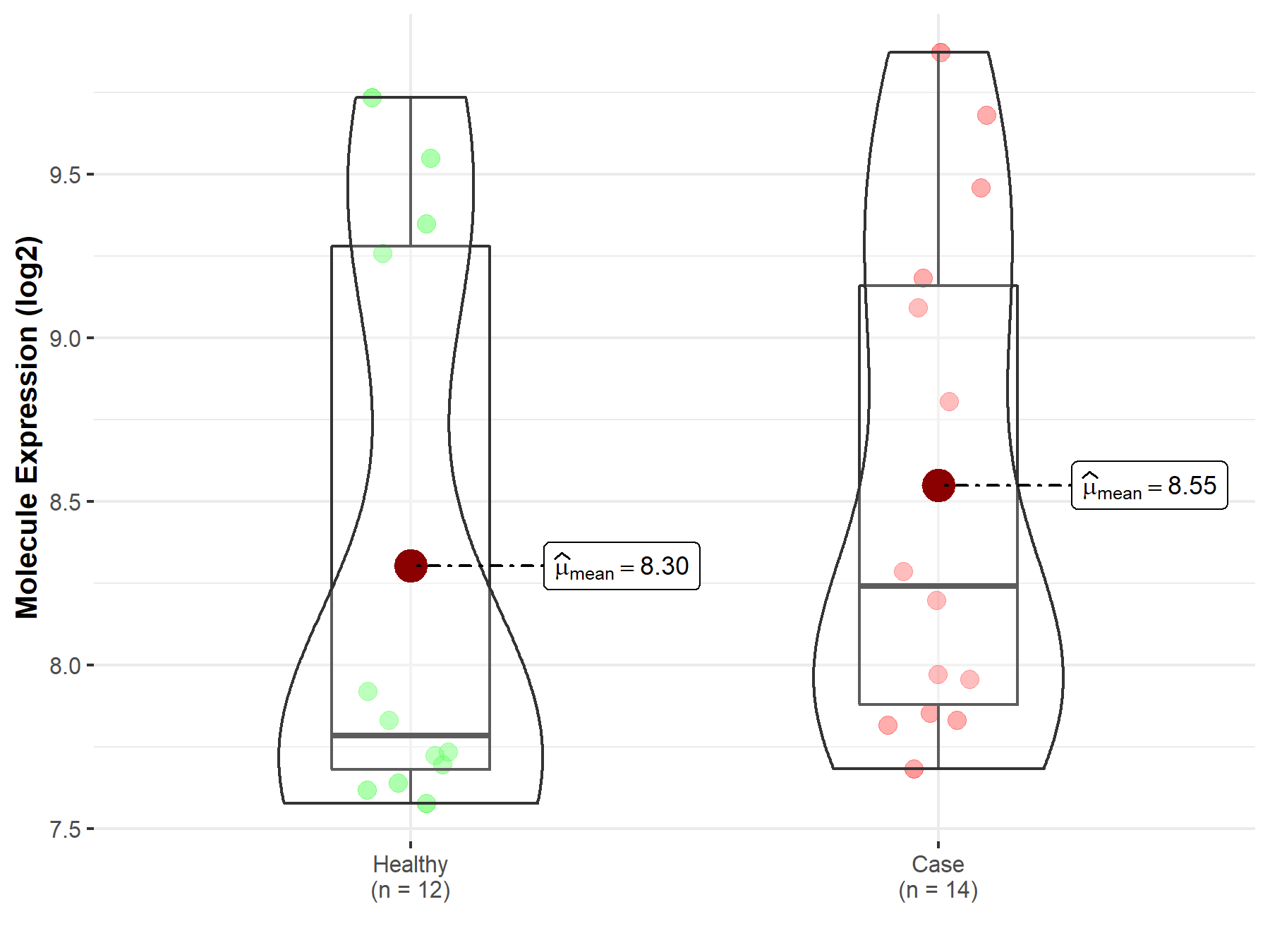
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

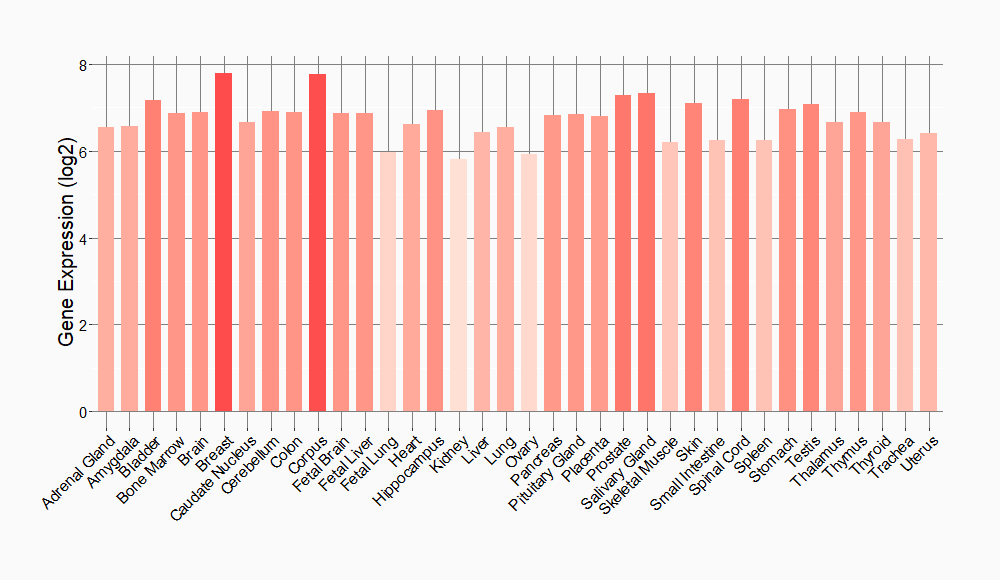
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
