Molecule Information
General Information of the Molecule (ID: Mol00241)
| Name |
Serine-protein kinase ATM (ATM)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Ataxia telangiectasia mutated; A-T mutated
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATM
|
||||
| Gene ID | |||||
| Location |
chr11:108223044-108369102[+]
|
||||
| Sequence |
MSLVLNDLLICCRQLEHDRATERKKEVEKFKRLIRDPETIKHLDRHSDSKQGKYLNWDAV
FRFLQKYIQKETECLRIAKPNVSASTQASRQKKMQEISSLVKYFIKCANRRAPRLKCQEL LNYIMDTVKDSSNGAIYGADCSNILLKDILSVRKYWCEISQQQWLELFSVYFRLYLKPSQ DVHRVLVARIIHAVTKGCCSQTDGLNSKFLDFFSKAIQCARQEKSSSGLNHILAALTIFL KTLAVNFRIRVCELGDEILPTLLYIWTQHRLNDSLKEVIIELFQLQIYIHHPKGAKTQEK GAYESTKWRSILYNLYDLLVNEISHIGSRGKYSSGFRNIAVKENLIELMADICHQVFNED TRSLEISQSYTTTQRESSDYSVPCKRKKIELGWEVIKDHLQKSQNDFDLVPWLQIATQLI SKYPASLPNCELSPLLMILSQLLPQQRHGERTPYVLRCLTEVALCQDKRSNLESSQKSDL LKLWNKIWCITFRGISSEQIQAENFGLLGAIIQGSLVEVDREFWKLFTGSACRPSCPAVC CLTLALTTSIVPGTVKMGIEQNMCEVNRSFSLKESIMKWLLFYQLEGDLENSTEVPPILH SNFPHLVLEKILVSLTMKNCKAAMNFFQSVPECEHHQKDKEELSFSEVEELFLQTTFDKM DFLTIVRECGIEKHQSSIGFSVHQNLKESLDRCLLGLSEQLLNNYSSEITNSETLVRCSR LLVGVLGCYCYMGVIAEEEAYKSELFQKAKSLMQCAGESITLFKNKTNEEFRIGSLRNMM QLCTRCLSNCTKKSPNKIASGFFLRLLTSKLMNDIADICKSLASFIKKPFDRGEVESMED DTNGNLMEVEDQSSMNLFNDYPDSSVSDANEPGESQSTIGAINPLAEEYLSKQDLLFLDM LKFLCLCVTTAQTNTVSFRAADIRRKLLMLIDSSTLEPTKSLHLHMYLMLLKELPGEEYP LPMEDVLELLKPLSNVCSLYRRDQDVCKTILNHVLHVVKNLGQSNMDSENTRDAQGQFLT VIGAFWHLTKERKYIFSVRMALVNCLKTLLEADPYSKWAILNVMGKDFPVNEVFTQFLAD NHHQVRMLAAESINRLFQDTKGDSSRLLKALPLKLQQTAFENAYLKAQEGMREMSHSAEN PETLDEIYNRKSVLLTLIAVVLSCSPICEKQALFALCKSVKENGLEPHLVKKVLEKVSET FGYRRLEDFMASHLDYLVLEWLNLQDTEYNLSSFPFILLNYTNIEDFYRSCYKVLIPHLV IRSHFDEVKSIANQIQEDWKSLLTDCFPKILVNILPYFAYEGTRDSGMAQQRETATKVYD MLKSENLLGKQIDHLFISNLPEIVVELLMTLHEPANSSASQSTDLCDFSGDLDPAPNPPH FPSHVIKATFAYISNCHKTKLKSILEILSKSPDSYQKILLAICEQAAETNNVYKKHRILK IYHLFVSLLLKDIKSGLGGAWAFVLRDVIYTLIHYINQRPSCIMDVSLRSFSLCCDLLSQ VCQTAVTYCKDALENHLHVIVGTLIPLVYEQVEVQKQVLDLLKYLVIDNKDNENLYITIK LLDPFPDHVVFKDLRITQQKIKYSRGPFSLLEEINHFLSVSVYDALPLTRLEGLKDLRRQ LELHKDQMVDIMRASQDNPQDGIMVKLVVNLLQLSKMAINHTGEKEVLEAVGSCLGEVGP IDFSTIAIQHSKDASYTKALKLFEDKELQWTFIMLTYLNNTLVEDCVKVRSAAVTCLKNI LATKTGHSFWEIYKMTTDPMLAYLQPFRTSRKKFLEVPRFDKENPFEGLDDINLWIPLSE NHDIWIKTLTCAFLDSGGTKCEILQLLKPMCEVKTDFCQTVLPYLIHDILLQDTNESWRN LLSTHVQGFFTSCLRHFSQTSRSTTPANLDSESEHFFRCCLDKKSQRTMLAVVDYMRRQK RPSSGTIFNDAFWLDLNYLEVAKVAQSCAAHFTALLYAEIYADKKSMDDQEKRSLAFEEG SQSTTISSLSEKSKEETGISLQDLLLEIYRSIGEPDSLYGCGGGKMLQPITRLRTYEHEA MWGKALVTYDLETAIPSSTRQAGIIQALQNLGLCHILSVYLKGLDYENKDWCPELEELHY QAAWRNMQWDHCTSVSKEVEGTSYHESLYNALQSLRDREFSTFYESLKYARVKEVEEMCK RSLESVYSLYPTLSRLQAIGELESIGELFSRSVTHRQLSEVYIKWQKHSQLLKDSDFSFQ EPIMALRTVILEILMEKEMDNSQRECIKDILTKHLVELSILARTFKNTQLPERAIFQIKQ YNSVSCGVSEWQLEEAQVFWAKKEQSLALSILKQMIKKLDASCAANNPSLKLTYTECLRV CGNWLAETCLENPAVIMQTYLEKAVEVAGNYDGESSDELRNGKMKAFLSLARFSDTQYQR IENYMKSSEFENKQALLKRAKEEVGLLREHKIQTNRYTVKVQRELELDELALRALKEDRK RFLCKAVENYINCLLSGEEHDMWVFRLCSLWLENSGVSEVNGMMKRDGMKIPTYKFLPLM YQLAARMGTKMMGGLGFHEVLNNLISRISMDHPHHTLFIILALANANRDEFLTKPEVARR SRITKNVPKQSSQLDEDRTEAANRIICTIRSRRPQMVRSVEALCDAYIILANLDATQWKT QRKGINIPADQPITKLKNLEDVVVPTMEIKVDHTGEYGNLVTIQSFKAEFRLAGGVNLPK IIDCVGSDGKERRQLVKGRDDLRQDAVMQQVFQMCNTLLQRNTETRKRKLTICTYKVVPL SQRSGVLEWCTGTVPIGEFLVNNEDGAHKRYRPNDFSAFQCQKKMMEVQKKSFEEKYEVF MDVCQNFQPVFRYFCMEKFLDPAIWFEKRLAYTRSVATSSIVGYILGLGDRHVQNILINE QSAELVHIDLGVAFEQGKILPTPETVPFRLTRDIVDGMGITGVEGVFRRCCEKTMEVMRN SQETLLTIVEVLLYDPLFDWTMNPLKALYLQQRPEDETELHPTLNADDQECKRNLSDIDQ SFNKVAERVLMRLQEKLKGVEEGTVLSVGGQVNLLIQQAIDPKNLSRLFPGWKAWV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Serine/threonine protein kinase which activates checkpoint signaling upon double strand breaks (DSBs), apoptosis and genotoxic stresses such as ionizing ultraviolet A light (UVA), thereby acting as a DNA damage sensor. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates 'Ser-139' of histone variant H2AX at double strand breaks (DSBs), thereby regulating DNA damage response mechanism. Also plays a role in pre-B cell allelic exclusion, a process leading to expression of a single immunoglobulin heavy chain allele to enforce clonality and monospecific recognition by the B-cell antigen receptor (BCR) expressed on individual B-lymphocytes. After the introduction of DNA breaks by the RAG complex on one immunoglobulin allele, acts by mediating a repositioning of the second allele to pericentromeric heterochromatin, preventing accessibility to the RAG complex and recombination of the second allele. Also involved in signal transduction and cell cycle control. May function as a tumor suppressor. Necessary for activation of ABL1 and SAPK. Phosphorylates DYRK2, CHEK2, p53/TP53, FBXW7, FANCD2, NFKBIA, BRCA1, CTIP, nibrin (NBN), TERF1, UFL1, RAD9, UBQLN4 and DCLRE1C. May play a role in vesicle and/or protein transport. Could play a role in T-cell development, gonad and neurological function. Plays a role in replication-dependent histone mRNA degradation. Binds DNA ends. Phosphorylation of DYRK2 in nucleus in response to genotoxic stress prevents its MDM2-mediated ubiquitination and subsequent proteasome degradation. Phosphorylates ATF2 which stimulates its function in DNA damage response. Phosphorylates ERCC6 which is essential for its chromatin remodeling activity at DNA double-strand breaks. Phosphorylates TTC5/STRAP at 'Ser-203' in the cytoplasm in response to DNA damage, which promotes TTC5/STRAP nuclear localization.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
5 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [1] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Fludarabine | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Inhibition | hsa04218 | |
| Experiment for Molecule Alteration |
Next-generation sequencing assay | |||
| Mechanism Description | Genes belonging to the DNA damage response and cell cycle control (TP53, ATM, POT1, BIRC3) happen to be more frequently mutated in uCLL cases. However, DNA-damaging chemotherapy results in the development of chemo-resistance in most of the cases, which has been initially attributed to the selection of driver mutations affecting genes of the DNA-damage response pathways, such as TP53 and ATM. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Mature B-cell neoplasms [.] | [2] | |||
| Resistant Disease | Mature B-cell neoplasms [.] | |||
| Resistant Drug | Inotuzumab ozogamicin | |||
| Molecule Alteration | Missense mutation | Methylation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | pre-InO and/or post-InO tumor cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
GeneSeq assay; Mutation assay | |||
| Mechanism Description | Multiple mechanisms drive CD22 antigen escape, including epitope loss (protein truncation and destabilization) and epitope alteration.Hypermutation caused by error-prone DNA damage repair may serve as a driver of CD22 mutation and escape. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [3] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | We validated ATM as a bona fide target of miR-203 in CRC cells. Mutation of the putative miR-203 binding site in the 3' untranslated region (3'UTR) of the ATM mRNA abolished the inhibitory effect of miR-203 on ATM. Furthermore, stable knockdown of ATM induced resistance to oxaliplatin in chemo-na ve CRC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen | |||
| Molecule Alteration | Missense mutation | p.I2948F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.I2948F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen/Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.I2948F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.27E-43; Fold-change: 4.17E-01; Z-score: 1.18E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.59E-04; Fold-change: 2.16E-01; Z-score: 4.77E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
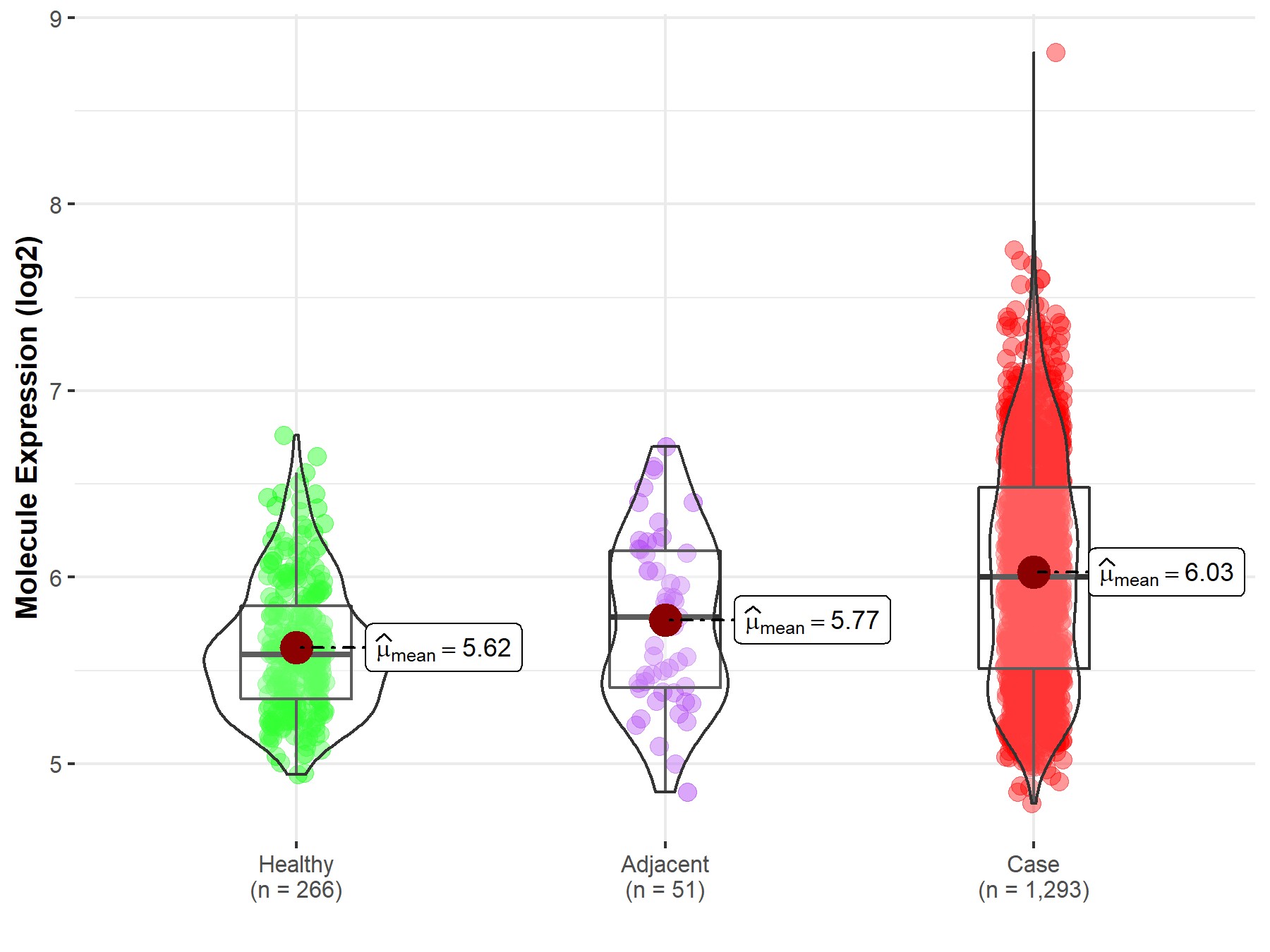
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

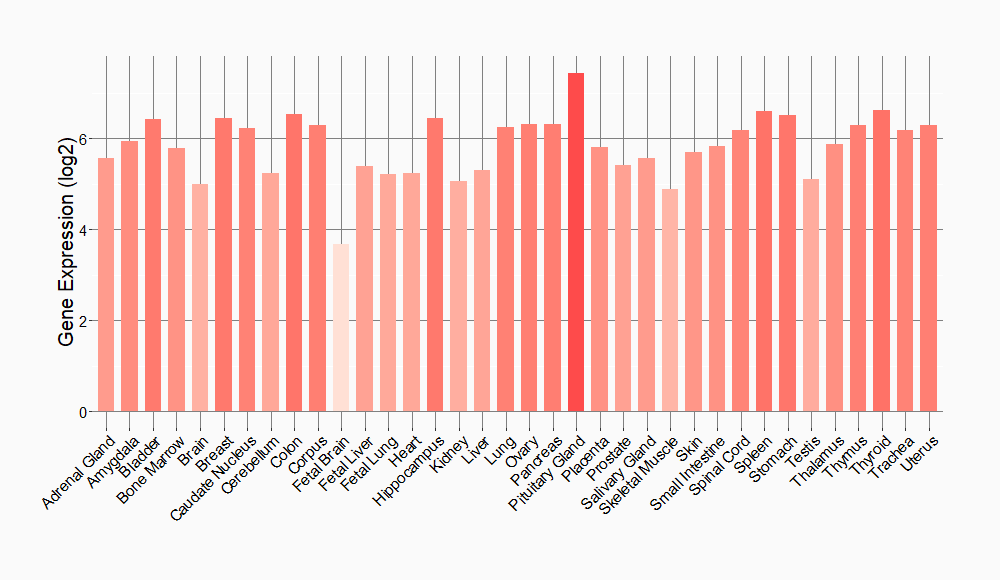
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
