Molecule Information
General Information of the Molecule (ID: Mol00202)
| Name |
Autophagy-related protein 16-1 (ATG16L1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
APG16-like 1; APG16L; UNQ9393/PRO34307
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATG16L1
|
||||
| Gene ID | |||||
| Location |
chr2:233210051-233295674[+]
|
||||
| Sequence |
MSSGLRAADFPRWKRHISEQLRRRDRLQRQAFEEIILQYNKLLEKSDLHSVLAQKLQAEK
HDVPNRHEISPGHDGTWNDNQLQEMAQLRIKHQEELTELHKKRGELAQLVIDLNNQMQRK DREMQMNEAKIAECLQTISDLETECLDLRTKLCDLERANQTLKDEYDALQITFTALEGKL RKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVEQDDDI EVIVDETSDHTEETSPVRAISRAATKRLSQPAGGLLDSITNIFGRRSVSSFPVPQDNVDT HPGSGKEVRVPATALCVFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWEVFGEKCEFKG SLSGSNAGITSIEFDSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDN ARIVSGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTEQCVMSGHFDKKIRFWDIRSES IVREMELLGKITALDLNPERTELLSCSRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRV VFSPDGSYVAAGSAEGSLYIWSVLTGKVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCK AVLWAQY Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Plays an essential role in both canonical and non-canonical autophagy: interacts with ATG12-ATG5 to mediate the lipidation to ATG8 family proteins (MAP1LC3A, MAP1LC3B, MAP1LC3C, GABARAPL1, GABARAPL2 and GABARAP). Acts as a molecular hub, coordinating autophagy pathways via distinct domains that support either canonical or non-canonical signaling. During canonical autophagy, interacts with ATG12-ATG5 to mediate the conjugation of phosphatidylethanolamine (PE) to ATG8 proteins, to produce a membrane-bound activated form of ATG8. Thereby, controls the elongation of the nascent autophagosomal membrane. Also involved in non-canonical autophagy, a parallel pathway involving conjugation of ATG8 proteins to single membranes at endolysosomal compartments, probably by catalyzing conjugation of phosphatidylserine (PS) to ATG8. Non-canonical autophagy plays a key role in epithelial cells to limit lethal infection by influenza A (IAV) virus. Regulates mitochondrial antiviral signaling (MAVS)-dependent type I interferon (IFN-I) production. Negatively regulates NOD1- and NOD2-driven inflammatory cytokine response. Instead, promotes with NOD2 an autophagy-dependent antibacterial pathway. Plays a role in regulating morphology and function of Paneth cell.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.81E-05 Fold-change: -9.87E-02 Z-score: -1.16E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR 874 could inhibit autophagy and sensitize GC cells to chemotherapy via the target gene ATG16L1. | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [2] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HFob 1.19 | Bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma, miR410 directly decreased ATG16L1 expression by targeting its 3'-untranslated region. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [2] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HFob 1.19 | Bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma, miR410 directly decreased ATG16L1 expression by targeting its 3'-untranslated region. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [2] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Sirolimus | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HFob 1.19 | Bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma, miR410 directly decreased ATG16L1 expression by targeting its 3'-untranslated region. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.81E-05; Fold-change: -6.17E-01; Z-score: -1.38E+01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.32E-02; Fold-change: -6.10E-01; Z-score: -1.26E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
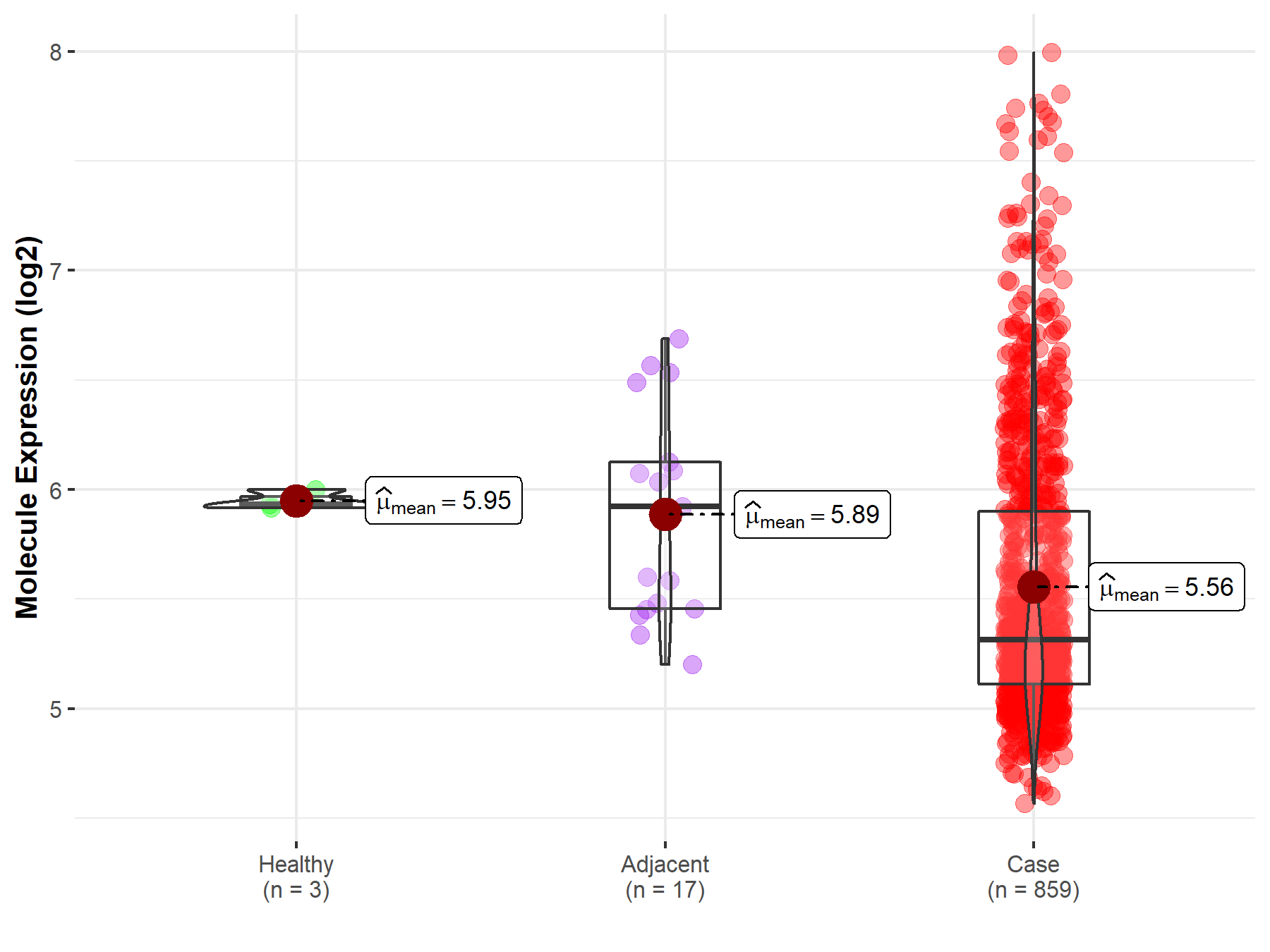
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

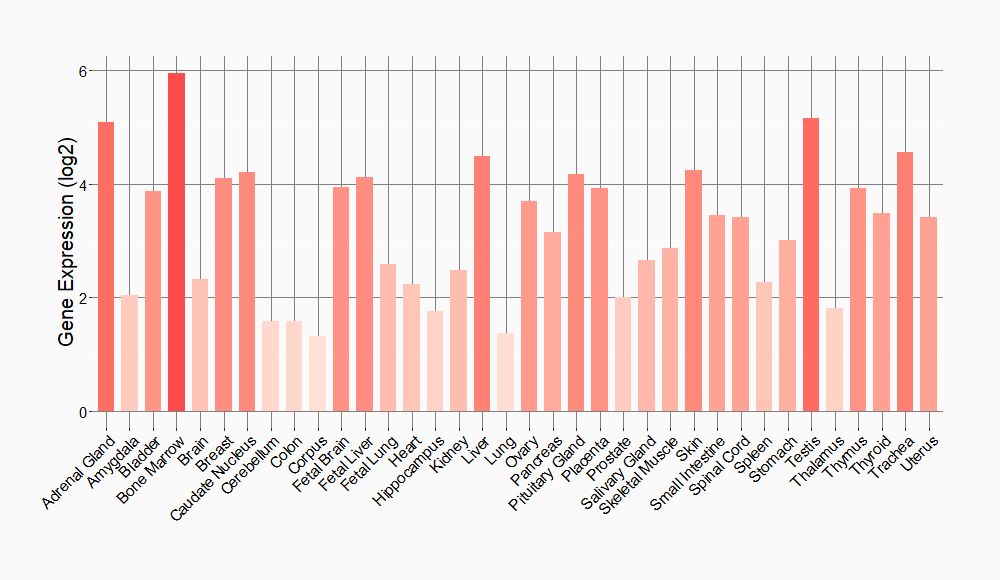
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
