Molecule Information
General Information of the Molecule (ID: Mol00181)
| Name |
Metalloproteinase inhibitor 3 (TIMP3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Protein MIG-5; Tissue inhibitor of metalloproteinases 3; TIMP-3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
TIMP3
|
||||
| Gene ID | |||||
| Location |
chr22:32801705-32863041[+]
|
||||
| Sequence |
MTPWLGLIVLLGSWSLGDWGAEACTCSPSHPQDAFCNSDIVIRAKVVGKKLVKEGPFGTL
VYTIKQMKMYRGFTKMPHVQYIHTEASESLCGLKLEVNKYQYLLTGRVYDGKMYTGLCNF VERWDQLTLSQRKGLNYRYHLGCNCKIKSCYYLPCFVTSKNECLWTDMLSNFGYPGYQSK HYACIRQKGGYCSWYRGWAPPDKSIINATDP Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Complexes with metalloproteinases (such as collagenases) and irreversibly inactivates them by binding to their catalytic zinc cofactor. May form part of a tissue-specific acute response to remodeling stimuli. Known to act on MMP-1, MMP-2, MMP-3, MMP-7, MMP-9, MMP-13, MMP-14 and MMP-15.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
7 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.70E-17 Fold-change: 1.51E+00 Z-score: 9.28E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-8 assay | |||
| Mechanism Description | Transfection of AS-miR-221 and AS-miR-222 dramatically inhibited expression of miR-221 and miR-222, respectively, in both MCF-7 and MDA-MB-231 cells (P<0.05-0.01). Down-regulation of miR-221/222 significantly increased the expression of TIMP3 compared with controls (P<0.05-0.01). The viability of estrogen receptor (ER)-positive MCF-7 cells transfected with AS-miR-221 or/and AS-miR-222 was significantly reduced by tamoxifen (P<0.05-0.01). Suppression of miRNA-221/222 increases the sensitivity of ER-positive MCF-7 breast cancer cells to tamoxifen. This effect is mediated through upregulation of TIMP3. These findings suggest that upregulation of TIMP3 via inhibition of miRNA-221/222 could be a promising therapeutic approach for breast cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [2] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-02 Fold-change: 2.68E+00 Z-score: 3.16E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [2] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-02 Fold-change: 2.68E+00 Z-score: 3.16E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [2] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-02 Fold-change: 2.68E+00 Z-score: 3.16E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [2] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Dovitinib lactate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-02 Fold-change: 2.68E+00 Z-score: 3.16E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Hepatocellular carcinoma | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.51E-20 Fold-change: -6.09E-01 Z-score: -9.89E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| TGF-beta signaling pathway | Activation | hsa04350 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | TIMP3 is a tumor suppressor and a validated miR-181 target. Ectopic expression and depletion of miR-181b showed that miR-181b enhanced MMP2 and MMP9 activity and promoted growth, clonogenic survival, migration and invasion of HCC cells that could be reversed by modulating TIMP3 level. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [4] | |||
| Sensitive Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Oral squamous cell carcinoma [ICD-11: 2B6E] | |||
| The Specified Disease | Oral cancer | |||
| The Studied Tissue | Oral tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.62E-01 Fold-change: 5.13E-02 Z-score: 1.43E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 |
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Annexin V-fluorescein isothiocyanate (FITC)/Hoechst double staining; MTT assay | |||
| Mechanism Description | OSCC cells are resistant to doxorubicin through upregulation of miR221, which in turn downregulates TIMP3. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [6] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Suppression of miR-21-5p expression sensitizes SGC7901/DOX cells to DOX via upregulating PTEN and TIMP3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [5] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Siha cells | Cervix uteri | Homo sapiens (Human) | CVCL_0032 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Colony formation assay | |||
| Mechanism Description | Down-regulation of LncRNA GAS5 can suppress TIMP3 and PDCD4 expression by enhancing miR-21 expression to suppress apoptosis and promote migration, invasion and cisplatin resistance in cervical cancer through the STAT3 signaling pathway. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Oral tissue | |
| The Specified Disease | Oral squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.62E-01; Fold-change: -2.10E-01; Z-score: -1.93E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.10E-16; Fold-change: 7.87E-01; Z-score: 1.38E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
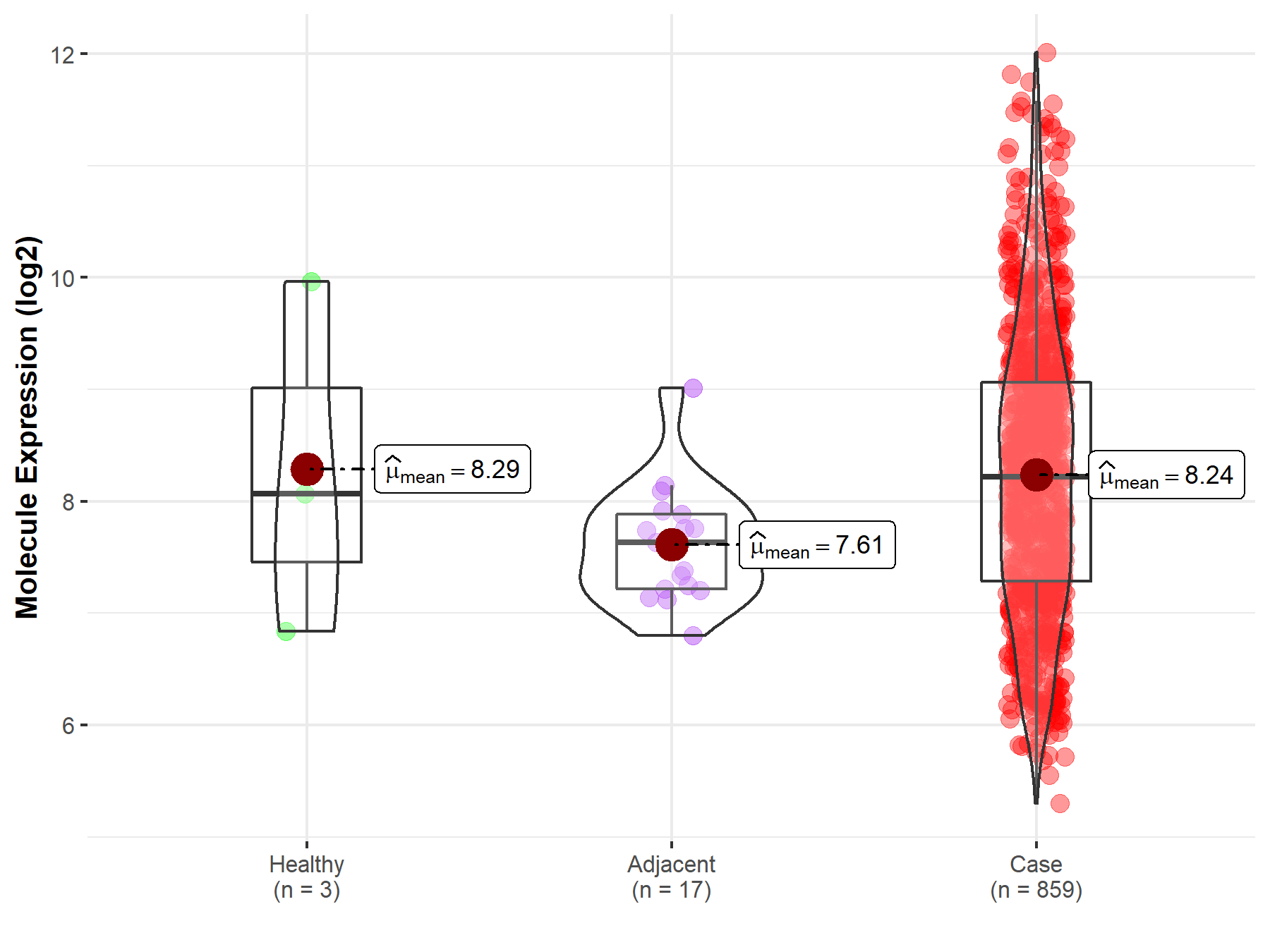
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.59E-01; Fold-change: 1.51E-01; Z-score: 9.60E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.55E-04; Fold-change: 5.85E-01; Z-score: 1.11E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
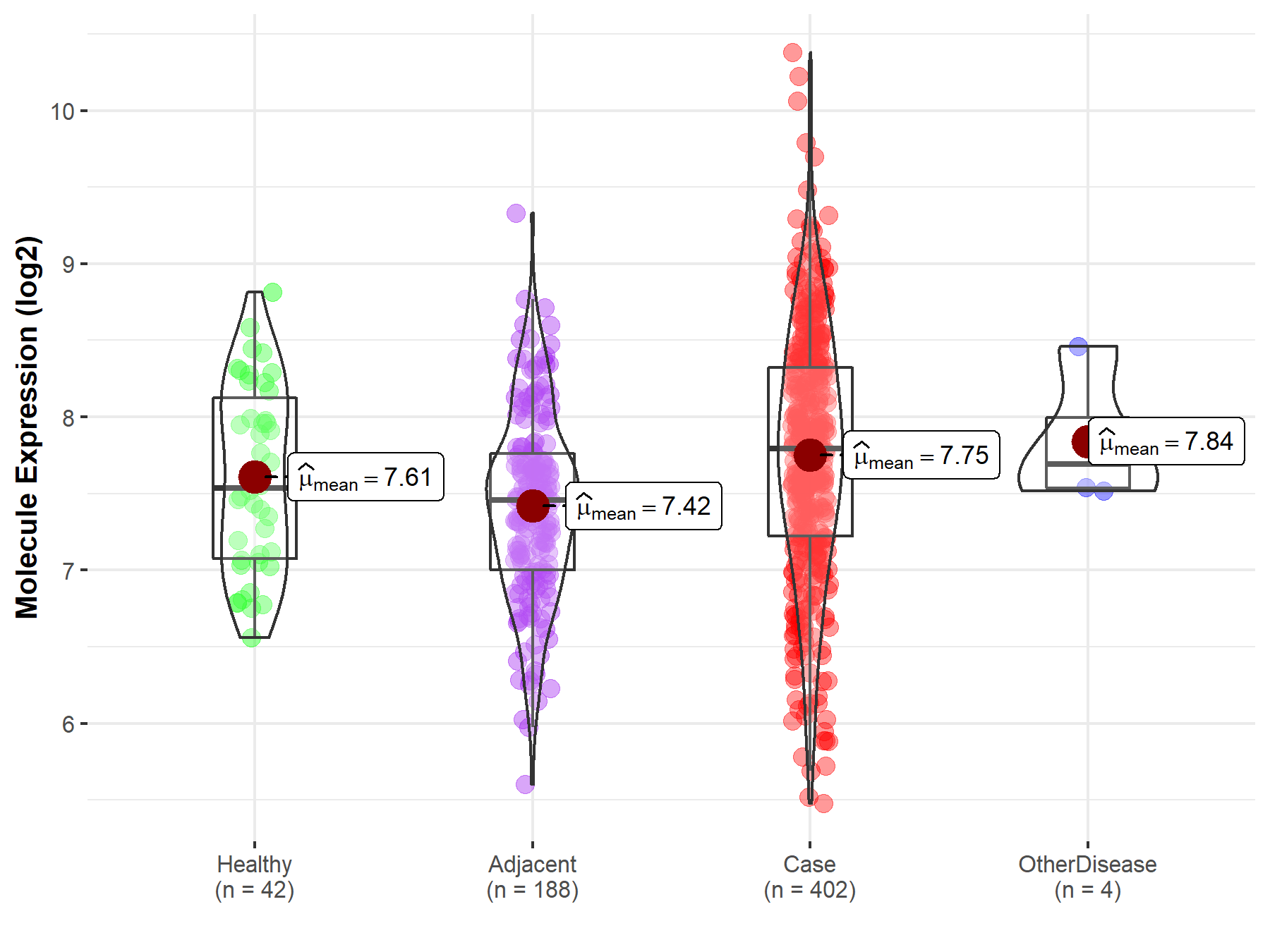
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.64E-01; Fold-change: 2.56E-01; Z-score: 4.24E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.21E-08; Fold-change: 3.37E-01; Z-score: 5.53E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 7.19E-01; Fold-change: 1.01E-01; Z-score: 2.30E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
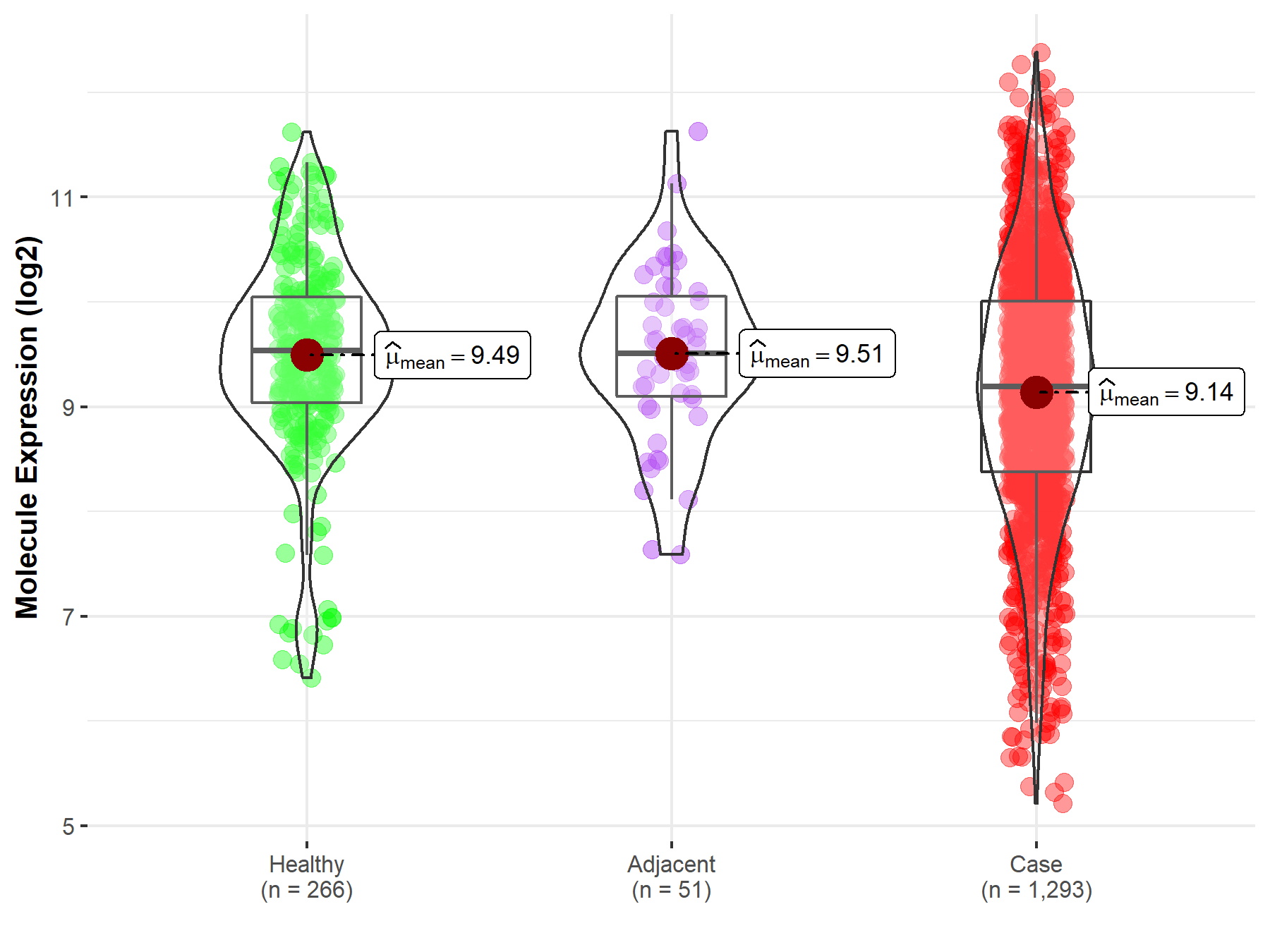
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.55E-07; Fold-change: -3.43E-01; Z-score: -3.64E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.24E-03; Fold-change: -3.17E-01; Z-score: -3.86E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
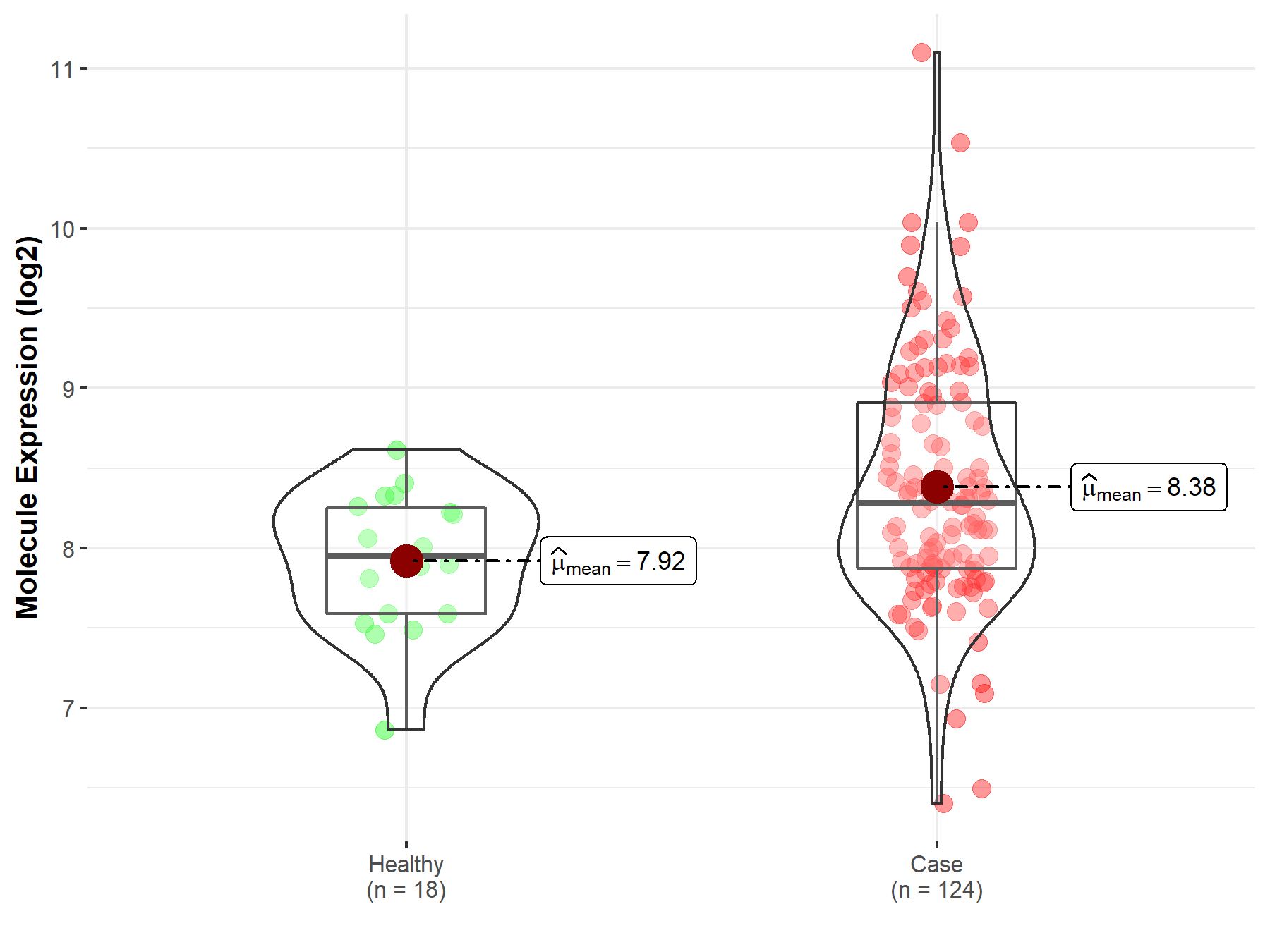
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.25E-04; Fold-change: 3.28E-01; Z-score: 7.48E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Kidney | |
| The Specified Disease | Kidney cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.26E-01; Fold-change: -4.20E-01; Z-score: -5.87E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.28E-07; Fold-change: -3.22E-01; Z-score: -5.62E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
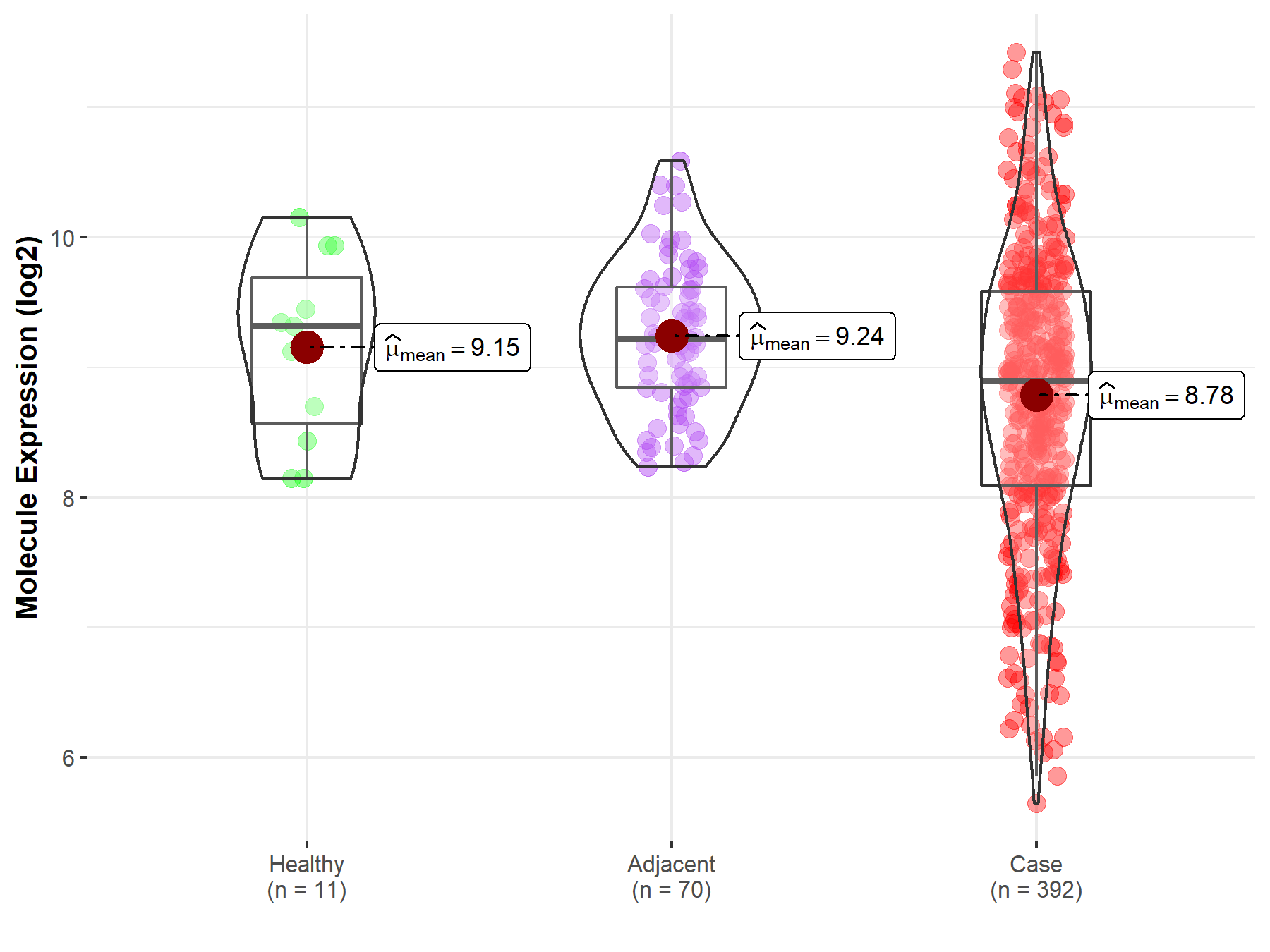
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
