Molecule Information
General Information of the Molecule (ID: Mol00165)
| Name |
Zinc finger protein SNAI1 (SNAI1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Protein snail homolog 1; Protein sna; SNAH
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
SNAI1
|
||||
| Gene ID | |||||
| Location |
chr20:49982980-49988886[+]
|
||||
| Sequence |
MPRSFLVRKPSDPNRKPNYSELQDSNPEFTFQQPYDQAHLLAAIPPPEILNPTASLPMLI
WDSVLAPQAQPIAWASLRLQESPRVAELTSLSDEDSGKGSQPPSPPSPAPSSFSSTSVSS LEAEAYAAFPGLGQVPKQLAQLSEAKDLQARKAFNCKYCNKEYLSLGALKMHIRSHTLPC VCGTCGKAFSRPWLLQGHVRTHTGEKPFSCPHCSRAFADRSNLRAHLQTHSDVKKYQCQA CARTFSRMSLLHKHQESGCSGCPR Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Involved in induction of the epithelial to mesenchymal transition (EMT), formation and maintenance of embryonic mesoderm, growth arrest, survival and cell migration. Binds to 3 E-boxes of the E-cadherin/CDH1 gene promoter and to the promoters of CLDN7 and KRT8 and, in association with histone demethylase KDM1A which it recruits to the promoters, causes a decrease in dimethylated H3K4 levels and represses transcription. The N-terminal SNAG domain competes with histone H3 for the same binding site on the histone demethylase complex formed by KDM1A and RCOR1, and thereby inhibits demethylation of histone H3 at 'Lys-4' (in vitro). During EMT, involved with LOXL2 in negatively regulating pericentromeric heterochromatin transcription. SNAI1 recruits LOXL2 to pericentromeric regions to oxidize histone H3 and repress transcription which leads to release of heterochromatin component CBX5/HP1A, enabling chromatin reorganization and acquisition of mesenchymal traits. Associates with EGR1 and SP1 to mediate tetradecanoyl phorbol acetate (TPA)-induced up-regulation of CDKN2B, possibly by binding to the CDKN2B promoter region 5'-TCACA-3. In addition, may also activate the CDKN2B promoter by itself.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
4 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [1] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.60E-02 Fold-change: 3.62E-02 Z-score: 2.09E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | A2780CP cells | Ovary | Homo sapiens (Human) | CVCL_0135 |
| A2780s cells | Ovary | Homo sapiens (Human) | CVCL_4863 | |
| C13 cells | Ovary | Homo sapiens (Human) | CVCL_0114 | |
| OV2008 cells | Ovary | Homo sapiens (Human) | CVCL_0473 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Snail overexpression could significantly attenuate miR-363-suppressed cisplatin resistance of EOC cells, suggesting that miR-363-regulated cisplatin resistance is mediated by snail-induced EMT in EOC cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [4] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| HO8910 cells | Ovary | Homo sapiens (Human) | CVCL_6868 | |
| CAOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0201 | |
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | miR30a/c-5p in turn directly inhibited DNMT1 as well as Snail. Forced expression of miR30a/c-5p or knocking down of DNMT1 and Snail promoted cisplatin susceptibility and partially reversed epithelial-mesenchymal transition (EMT) in CP70 cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [2] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.20E-01 Fold-change: -2.83E-02 Z-score: -1.27E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| SNAI1/IRS1/AKT signaling pathway | Regulation | N.A. | ||
| In Vitro Model | BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 |
| PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 | |
| SW1990 cells | Pancreas | Homo sapiens (Human) | CVCL_1723 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-30a overexpression suppresses cell proliferation, and sensitizes pancreatic cancer cells to gemcitabine and miR-30a overexpression reduced IRS1 and SNAI1 protein level. | |||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [5] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 |
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | |
| PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 | |
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | |
| AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 | |
| SW1990 cells | Pancreas | Homo sapiens (Human) | CVCL_1723 | |
| CFPAC1 cells | Pancreas | Homo sapiens (Human) | CVCL_1119 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Annexin-V/PI Apoptosis assay; TUNEL assay | |||
| Mechanism Description | miR153 enhanced gemcitabine sensitivity by targeting Snail in pancreatic cancer. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [3] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-04 Fold-change: -2.85E-02 Z-score: -3.91E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| miR125b/HER2/Snail1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Wound-healing assay; Transwell assay | |||
| Mechanism Description | TINCR, which is transcriptionally activated by H3k27 acetylation, upregulates HER-2 expression by downregulating miR-125b and TINCR promotes trastuzumab resistance-induced EMT by directly targeting Snail-1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [6] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay; Wound healing and transwell assay | |||
| Mechanism Description | Overexpression of miR647 sensitizes tumors to chemotherapy in vivo by reducing the expression levels of ANk2, FAk, MMP2, MMP12, CD44 and SNAIL1. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.29E-01; Fold-change: 2.40E-01; Z-score: 4.62E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.54E-06; Fold-change: 4.35E-01; Z-score: 1.69E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.20E-01; Fold-change: -1.09E-01; Z-score: -3.14E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.86E-03; Fold-change: -2.28E-01; Z-score: -5.65E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-04; Fold-change: -6.17E-02; Z-score: -1.47E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.77E-01; Fold-change: -1.83E-02; Z-score: -4.95E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.60E-02; Fold-change: 5.28E-02; Z-score: 2.85E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.29E-01; Fold-change: -1.40E-01; Z-score: -2.12E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
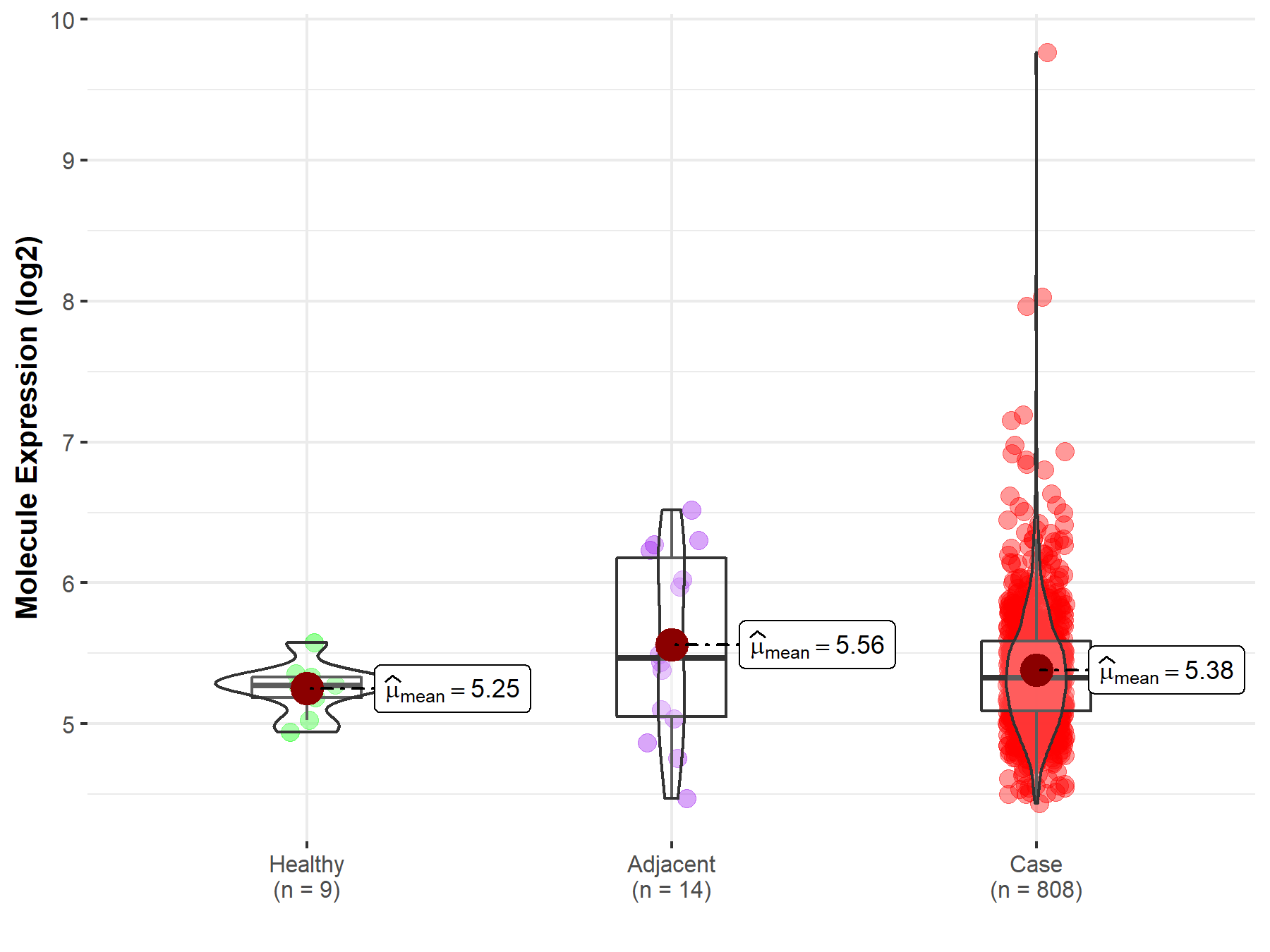
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
