Molecule Information
General Information of the Molecule (ID: Mol00135)
| Name |
phosphoinositide-3-dependent protein kinase 1 (PDPK1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
hPDK1; PDK1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PDPK1
|
||||
| Gene ID | |||||
| Location |
chr16:2537979-2603188[+]
|
||||
| Sequence |
MARTTSQLYDAVPIQSSVVLCSCPSPSMVRTQTESSTPPGIPGGSRQGPAMDGTAAEPRP
GAGSLQHAQPPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRHIIK ENKVPYVTRERDVMSRLDHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSFDET CTRFYTAEIVSALEYLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQARAN SFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVAGLPPFRAGNEYLIFQKIIKLEYD FPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFESVTWENLHQQTPPKLTA YLPAMSEDDEDCYGNYDNLLSQFGCMQVSSSSSSHSLSASDTGLPQRSGSNIEQYIHDLD SNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLLLTE GPHLYYVDPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCRKIQ EVWRQRYQSHPDAAVQ Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Serine/threonine kinase which acts as a master kinase, phosphorylating and activating a subgroup of the AGC family of protein kinases. Its targets include: protein kinase B (PKB/AKT1, PKB/AKT2, PKB/AKT3), p70 ribosomal protein S6 kinase (RPS6KB1), p90 ribosomal protein S6 kinase (RPS6KA1, RPS6KA2 and RPS6KA3), cyclic AMP-dependent protein kinase (PRKACA), protein kinase C (PRKCD and PRKCZ), serum and glucocorticoid-inducible kinase (SGK1, SGK2 and SGK3), p21-activated kinase-1 (PAK1), protein kinase PKN (PKN1 and PKN2). Plays a central role in the transduction of signals from insulin by providing the activating phosphorylation to PKB/AKT1, thus propagating the signal to downstream targets controlling cell proliferation and survival, as well as glucose and amino acid uptake and storage. Negatively regulates the TGF-beta-induced signaling by: modulating the association of SMAD3 and SMAD7 with TGF-beta receptor, phosphorylating SMAD2, SMAD3, SMAD4 and SMAD7, preventing the nuclear translocation of SMAD3 and SMAD4 and the translocation of SMAD7 from the nucleus to the cytoplasm in response to TGF-beta. Activates PPARG transcriptional activity and promotes adipocyte differentiation. Activates the NF-kappa-B pathway via phosphorylation of IKKB. The tyrosine phosphorylated form is crucial for the regulation of focal adhesions by angiotensin II. Controls proliferation, survival, and growth of developing pancreatic cells. Participates in the regulation of Ca(2+) entry and Ca(2+)-activated K(+) channels of mast cells. Essential for the motility of vascular endothelial cells (ECs) and is involved in the regulation of their chemotaxis. Plays a critical role in cardiac homeostasis by serving as a dual effector for cell survival and beta-adrenergic response. Plays an important role during thymocyte development by regulating the expression of key nutrient receptors on the surface of pre-T cells and mediating Notch-induced cell growth and proliferative responses. Provides negative feedback inhibition to toll-like receptor-mediated NF-kappa-B activation in macrophages. Isoform 3 is catalytically inactive.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [1] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Dichloroacetate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Neuroectodermal tumor | |||
| The Studied Tissue | Brainstem tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.99E-03 Fold-change: 8.06E-02 Z-score: 3.94E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DBTRG cells | Brain | Homo sapiens (Human) | CVCL_1169 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Colorimetric SRB assay | |||
| Mechanism Description | The potential of miR-144 overexpression to reduce GB cell malignancy, both by decreasing Cell migration and invasion abilities and by sensitizing resistant tumor cells to chemotherapy, paving the way to a novel and more effective GB therapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Erlotinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.73E-09 Fold-change: 3.48E-02 Z-score: 5.92E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [3] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SAS cells | Oral | Homo sapiens (Human) | CVCL_1675 |
| TW2.6 cells | Mouth | Homo sapiens (Human) | CVCL_GZ05 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Apoptosis rate assay | |||
| Mechanism Description | Immunohistochemical analysis revealed that higher PDK1 expression is associated with a poor prognosis in OSCC. The immunoprecipitation assay indicated PDK1/CD47 binding. PDK1 ligation significantly impaired OSCC orosphere formation and downregulated Sox2, Oct4, and CD133 expression. The combination of BX795 and cisplatin markedly reduced in OSCC cell's epithelial-mesenchymal transition, implying its synergistic effect. p-PDK1, CD47, Akt, PFKP, PDK3 and LDHA protein expression were significantly reduced, with the strongest inhibition in the combination group. Chemo/radiotherapy together with abrogation of PDK1 inhibits the oncogenic (Akt/CD47) and glycolytic (LDHA/PFKP/PDK3) signaling and, enhanced or sensitizes OSCC to the anticancer drug effect through inducing apoptosis and DNA damage together with metabolic reprogramming. | |||
| Disease Class: Head and neck cancer [ICD-11: 2D42.0] | [4] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Head and neck cancer [ICD-11: 2D42.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HNC SAS cells | Head and Neck | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | After PDK1 and PDK2 knockdown, we discovered increased ATP production and decreased lactate production in TGFbeta1-treated and untreated HNC cells. However, only PDK2 silencing significantly inhibited the clonogenic ability of HNC cells. We subsequently found that TGFbeta1-promoted migration and invasion capabilities were decreased in PDK1 and PDK2 knockdown cells. The tumor spheroid-forming capability, motility, CSC genes, and multidrug-resistant genes were downregulated in PDK1 and PDK2 silencing CSCs. PDK1 and PDK2 inhibition reversed cisplatin and gemcitabine resistance of CSCs, but not paclitaxel resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Head and neck cancer [ICD-11: 2D42.0] | [4] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Head and neck cancer [ICD-11: 2D42.0] | |||
| Resistant Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HNC SAS cells | Head and Neck | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | After PDK1 and PDK2 knockdown, we discovered increased ATP production and decreased lactate production in TGFbeta1-treated and untreated HNC cells. However, only PDK2 silencing significantly inhibited the clonogenic ability of HNC cells. We subsequently found that TGFbeta1-promoted migration and invasion capabilities were decreased in PDK1 and PDK2 knockdown cells. The tumor spheroid-forming capability, motility, CSC genes, and multidrug-resistant genes were downregulated in PDK1 and PDK2 silencing CSCs. PDK1 and PDK2 inhibition reversed cisplatin and gemcitabine resistance of CSCs, but not paclitaxel resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Icotinib hydrochloride | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | |||
Investigative Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [3] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Resistant Drug | BX795 | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SAS cells | Oral | Homo sapiens (Human) | CVCL_1675 |
| TW2.6 cells | Mouth | Homo sapiens (Human) | CVCL_GZ05 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Apoptosis rate assay | |||
| Mechanism Description | Immunohistochemical analysis revealed that higher PDK1 expression is associated with a poor prognosis in OSCC. The immunoprecipitation assay indicated PDK1/CD47 binding. PDK1 ligation significantly impaired OSCC orosphere formation and downregulated Sox2, Oct4, and CD133 expression. The combination of BX795 and cisplatin markedly reduced in OSCC cell's epithelial-mesenchymal transition, implying its synergistic effect. p-PDK1, CD47, Akt, PFKP, PDK3 and LDHA protein expression were significantly reduced, with the strongest inhibition in the combination group. Chemo/radiotherapy together with abrogation of PDK1 inhibits the oncogenic (Akt/CD47) and glycolytic (LDHA/PFKP/PDK3) signaling and, enhanced or sensitizes OSCC to the anticancer drug effect through inducing apoptosis and DNA damage together with metabolic reprogramming. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [5] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | PI3K pathway inhibitors | |||
| Molecule Alteration | Copy number gain | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Breast | N.A. | ||
| In Vivo Model | SCID/NCr (BALB/c background) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Mechanism Description | The copy number gain in gene PDPK1 cause the sensitivity of PI3K pathway inhibitors by unusual activation of pro-survival pathway. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.27E-130; Fold-change: -5.10E-01; Z-score: -1.73E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.26E-02; Fold-change: -2.56E-01; Z-score: -3.17E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
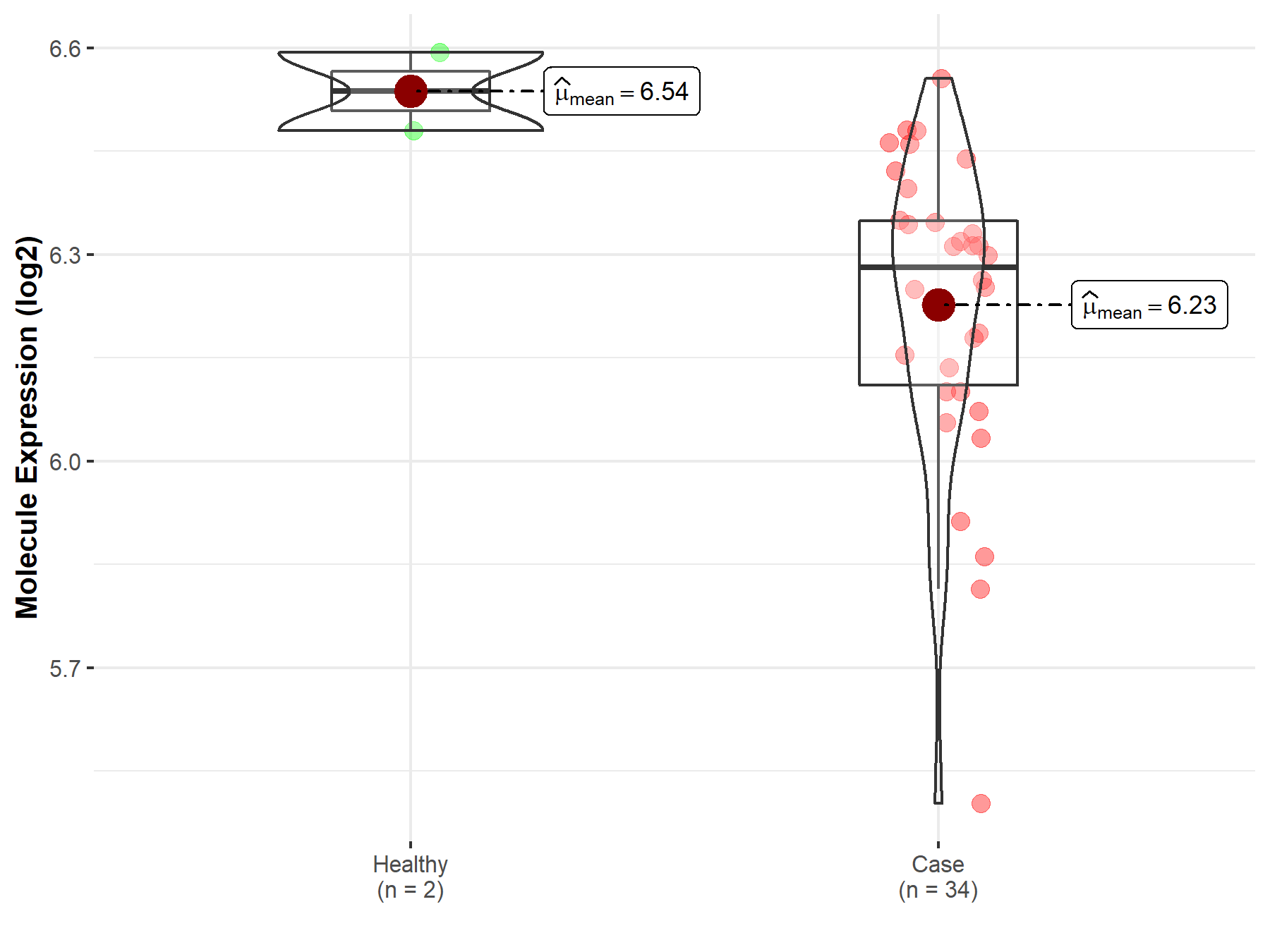
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.77E-01; Fold-change: 1.36E-01; Z-score: 7.98E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.99E-03; Fold-change: 2.98E-01; Z-score: 9.97E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.73E-09; Fold-change: 2.51E-01; Z-score: 8.32E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.21E-06; Fold-change: 1.84E-01; Z-score: 3.88E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.41E-01; Fold-change: 2.00E-02; Z-score: 6.33E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.13E-05; Fold-change: 5.85E-01; Z-score: 1.14E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

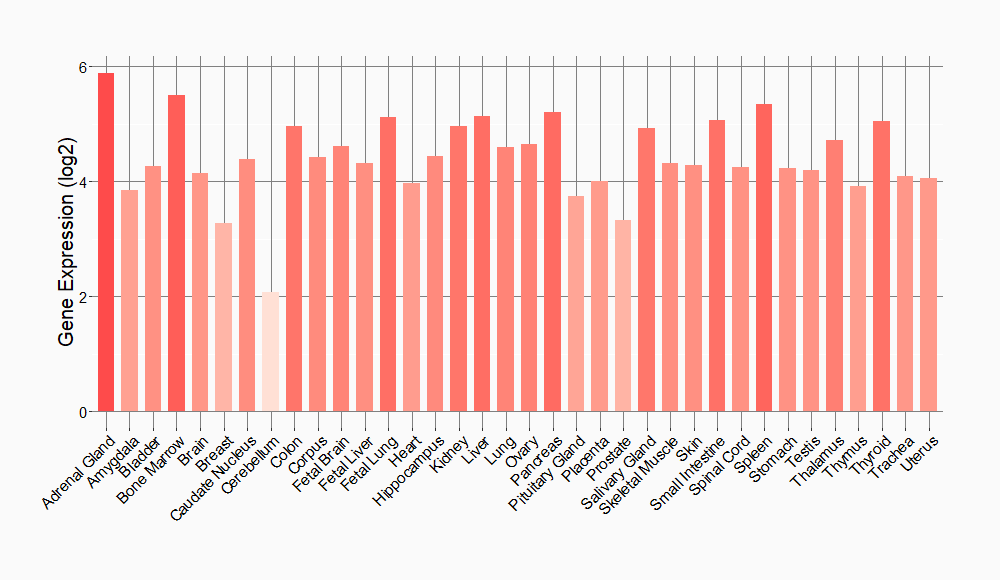
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
