Molecule Information
General Information of the Molecule (ID: Mol00115)
| Name |
Autophagy-related protein LC3 B (MAP1LC3B)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Autophagy-related protein LC3 B; Autophagy-related ubiquitin-like modifier LC3 B; MAP1 light chain 3-like protein 2; MAP1A/MAP1B light chain 3 B; MAP1A/MAP1B LC3 B; Microtubule-associated protein 1 light chain 3 beta; MAP1ALC3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
MAP1LC3B
|
||||
| Gene ID | |||||
| Location |
chr16:87383953-87404779[+]
|
||||
| Sequence |
MPSEKTFKQRRTFEQRVEDVRLIREQHPTKIPVIIERYKGEKQLPVLDKTKFLVPDHVNM
SELIKIIRRRLQLNANQAFFLLVNGHSMVSVSTPISEVYESEKDEDGFLYMVYASQETFG MKLSV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Ubiquitin-like modifier involved in formation of autophagosomal vacuoles (autophagosomes). Plays a role in mitophagy which contributes to regulate mitochondrial quantity and quality by eliminating the mitochondria to a basal level to fulfill cellular energy requirements and preventing excess ROS production. In response to cellular stress and upon mitochondria fission, binds C-18 ceramides and anchors autophagolysosomes to outer mitochondrial membranes to eliminate damaged mitochondria. While LC3s are involved in elongation of the phagophore membrane, the GABARAP/GATE-16 subfamily is essential for a later stage in autophagosome maturation. Promotes primary ciliogenesis by removing OFD1 from centriolar satellites via the autophagic pathway. Through its interaction with the reticulophagy receptor TEX264, participates in the remodeling of subdomains of the endoplasmic reticulum into autophagosomes upon nutrient stress, which then fuse with lysosomes for endoplasmic reticulum turnover. Upon nutrient stress, directly recruits cofactor JMY to the phagophore membrane surfaces and promotes JMY's actin nucleation activity and autophagosome biogenesis during autophagy.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [1] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | Penfluridol | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.86E-03 Fold-change: 9.00E-02 Z-score: 3.21E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A549/Taxol cells | Lung | Homo sapiens (Human) | CVCL_W218 |
| SW403 cells | Colon | Homo sapiens (Human) | CVCL_0545 | |
| In Vivo Model | Athymic nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
SRB assay | |||
| Mechanism Description | One of the hallmarks of autophagy is the accumulation of LC3B and its localization in vesicular structures. We observed that penfluridol treatment enhanced the expression of LC3B and hence induced autophagy in pancreatic cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [2] | |||
| Sensitive Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell autophagy | Inhibition | hsa04140 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | CAL-27 cells | Tongue | Homo sapiens (Human) | CVCL_1107 |
| KB cells | Gastric | Homo sapiens (Human) | CVCL_0372 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | After HOTAIR silence, autophagy was inhibited with the downregulated expression of MAP1LC3B (microtubule-associated protein 1 light chain 3B), beclin1, and autophagy-related gene (ATG) 3 and ATG7. The expressions of mTOR increased, which promoted the sensitivity to cisplatin. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Oral tissue | |
| The Specified Disease | Oral squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.13E-02; Fold-change: 5.20E-01; Z-score: 7.53E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.44E-01; Fold-change: 1.72E-01; Z-score: 5.04E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

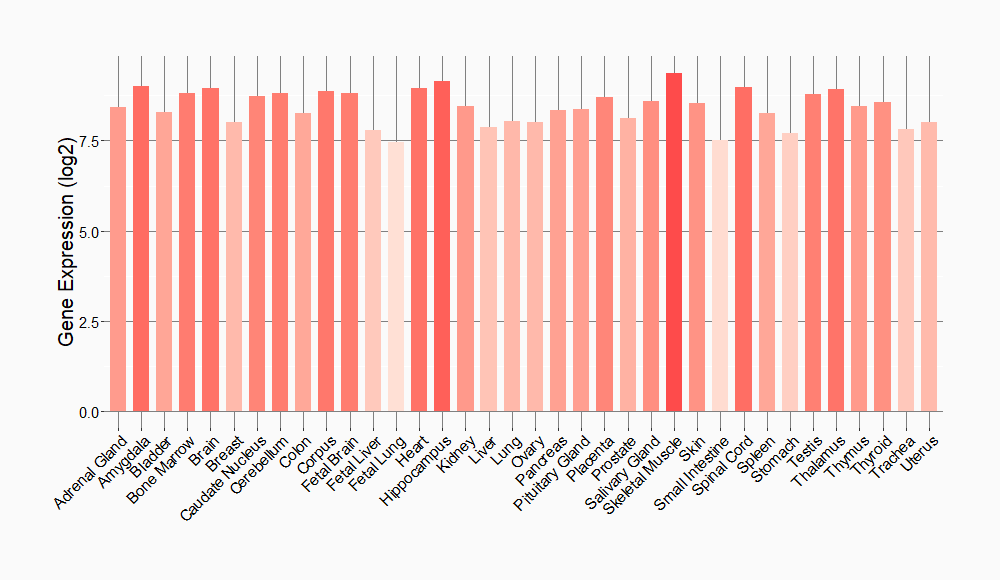
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
