Molecule Information
General Information of the Molecule (ID: Mol00096)
| Name |
Interferon alpha-inducible protein 27 (IFI27)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
p27; Interferon alpha-induced 11.5 kDa protein; Interferon-stimulated gene 12a protein; ISG12(a); ISG12A
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
IFI27
|
||||
| Gene ID | |||||
| Location |
chr14:94104836-94116695[+]
|
||||
| Sequence |
MEASALTSSAVTSVAKVVRVASGSAVVLPLARIATVVIGGVVAMAAVPMVLSAMGFTAAG
IASSSIAAKMMSAAAIANGGGVASGSLVATLQSLGATGLSGLTKFILGSIGSAIAAVIAR FY Click to Show/Hide
|
||||
| Function |
Probable adapter protein involved in different biological processes. Part of the signaling pathways that lead to apoptosis. Involved in type-I interferon-induced apoptosis characterized by a rapid and robust release of cytochrome C from the mitochondria and activation of BAX and caspases , and 9. Also functions in TNFSF10-induced apoptosis. May also have a function in the nucleus, where it may be involved in the interferon-induced negative regulation of the transcriptional activity of NR4A1, NR4A2 and NR4A3 through the enhancement of XPO1-mediated nuclear export of these nuclear receptors. May thereby play a role in the vascular response to injury. In the innate immune response, has an antiviral activity towards hepatitis C virus/HCV. May prevent the replication of the virus by recruiting both the hepatitis C virus non-structural protein 5A/NS5A and the ubiquitination machinery via SKP2, promoting the ubiquitin-mediated proteasomal degradation of NS5A.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | TRAIL | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Hepatocellular carcinoma | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.37E-02 Fold-change: -1.60E-01 Z-score: -1.68E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HLCZ01 cells | Hepatoma | Homo sapiens (Human) | CVCL_1J92 | |
| LH86 cells | Hepatoma | Homo sapiens (Human) | CVCL_8889 | |
| HLCZ02 cells | Liver | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-942 is upregulated in TRAIL-resistant cancer cells and decreased in TRAIL-sensitive ones. miR-942 is inversely correlated with ISG12a expression in cancer tissues and cells. AkT control TRAIL resistance of cancer cells through downregulation of ISG12a by miR-942. Down-regulation of ISG12a by miR-942 is needed to maintain the TRAIL-resistant phenotype. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | TRAIL | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | HGC27 cells | Gastric | Homo sapiens (Human) | CVCL_1279 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-942 is upregulated in TRAIL-resistant cancer cells and decreased in TRAIL-sensitive ones. miR-942 is inversely correlated with ISG12a expression in cancer tissues and cells. AkT control TRAIL resistance of cancer cells through downregulation of ISG12a by miR-942. Down-regulation of ISG12a by miR-942 is needed to maintain the TRAIL-resistant phenotype. | |||
| Disease Class: Human papillomavirus-related endocervical adenocarcinoma [ICD-11: 2C77.2] | [1] | |||
| Resistant Disease | Human papillomavirus-related endocervical adenocarcinoma [ICD-11: 2C77.2] | |||
| Resistant Drug | TRAIL | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-942 is upregulated in TRAIL-resistant cancer cells and decreased in TRAIL-sensitive ones. miR-942 is inversely correlated with ISG12a expression in cancer tissues and cells. AkT control TRAIL resistance of cancer cells through downregulation of ISG12a by miR-942. Down-regulation of ISG12a by miR-942 is needed to maintain the TRAIL-resistant phenotype. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.07E-01; Fold-change: 9.46E-01; Z-score: 6.24E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.05E-06; Fold-change: -6.65E-01; Z-score: -1.28E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.16E-09; Fold-change: 1.30E+00; Z-score: 1.22E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.00E-01; Fold-change: 3.17E-01; Z-score: 1.82E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 1.53E-01; Fold-change: -4.64E-01; Z-score: -9.07E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
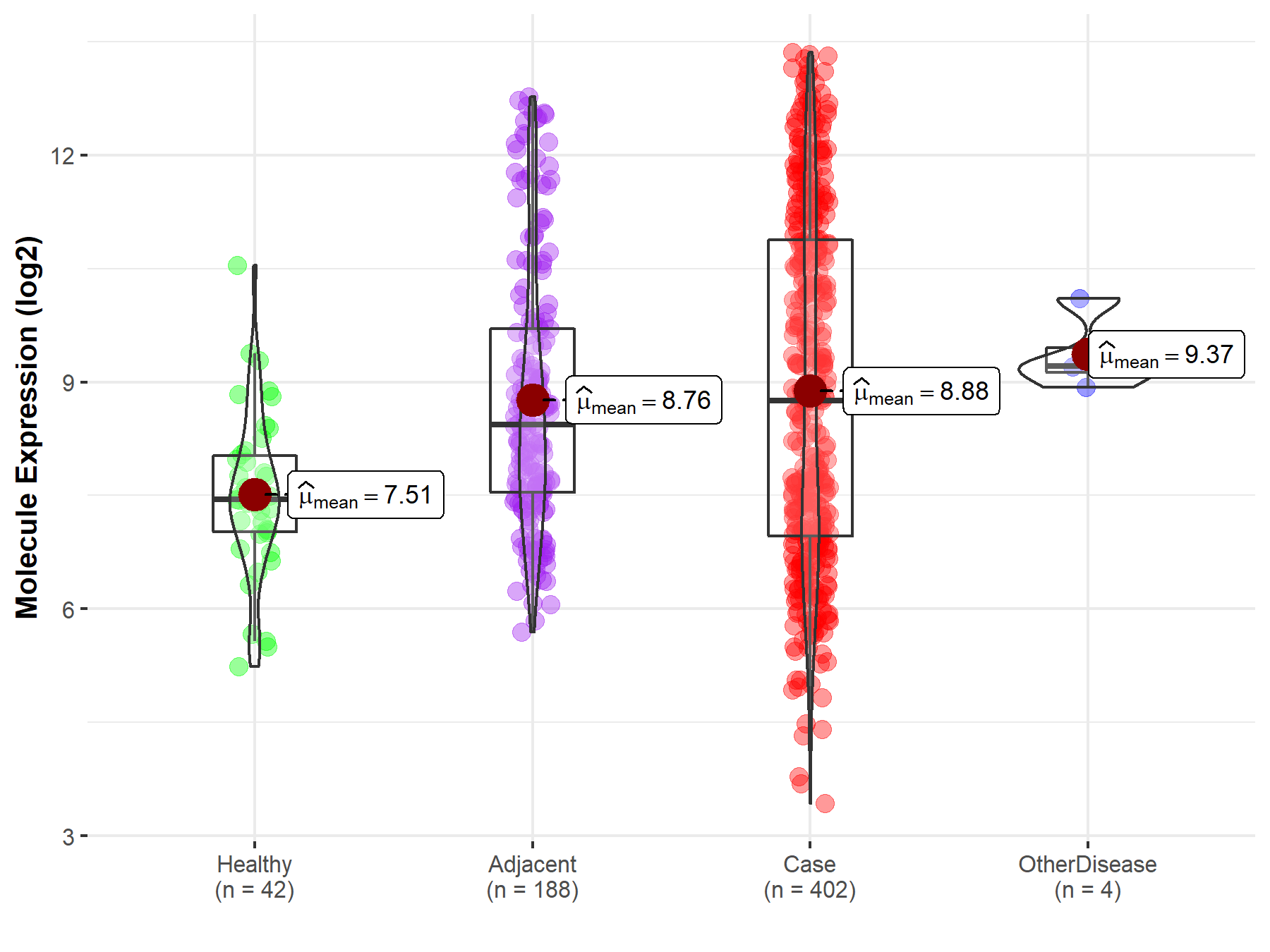
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.85E-02; Fold-change: 4.38E-01; Z-score: 4.37E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
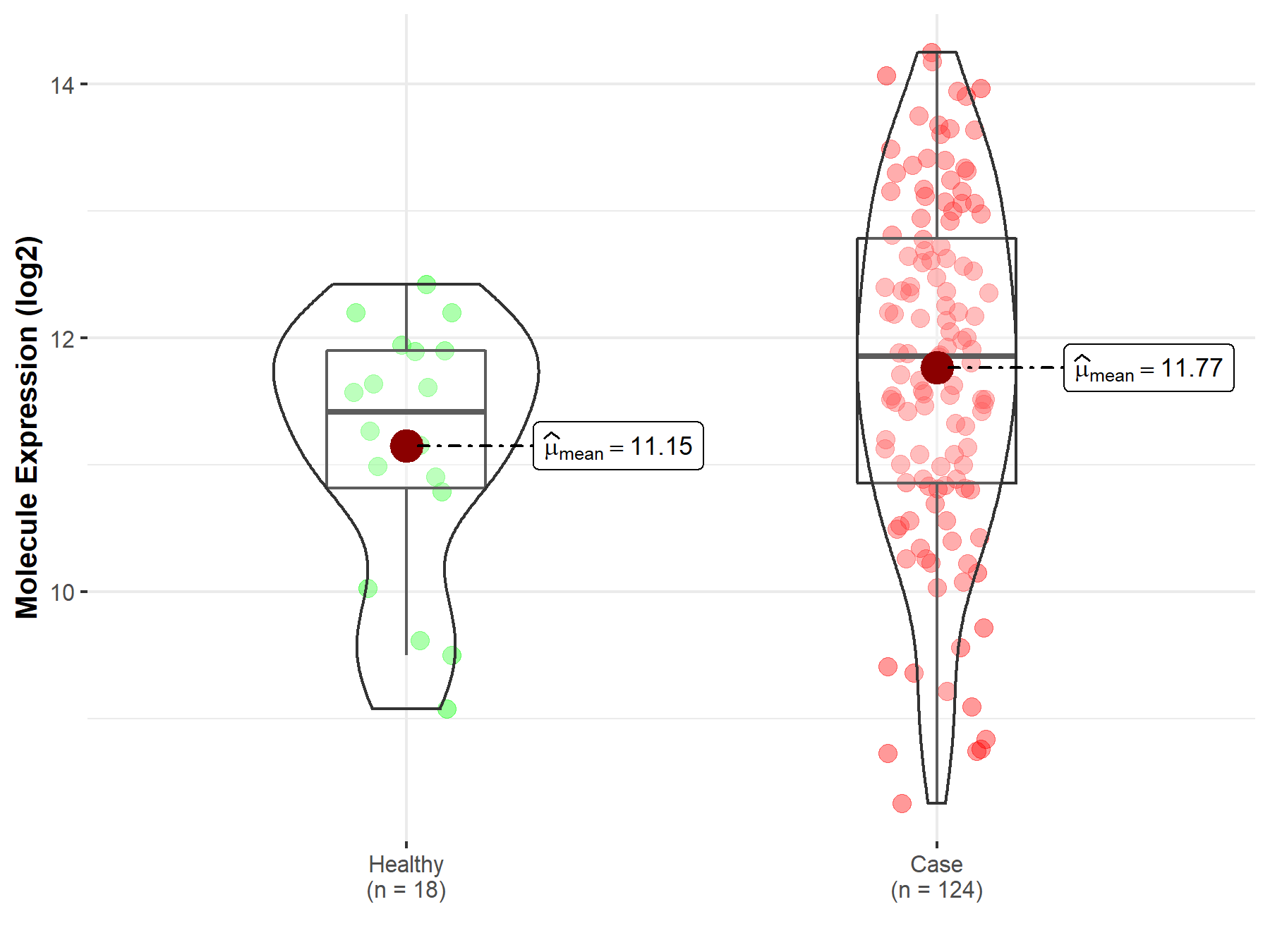
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

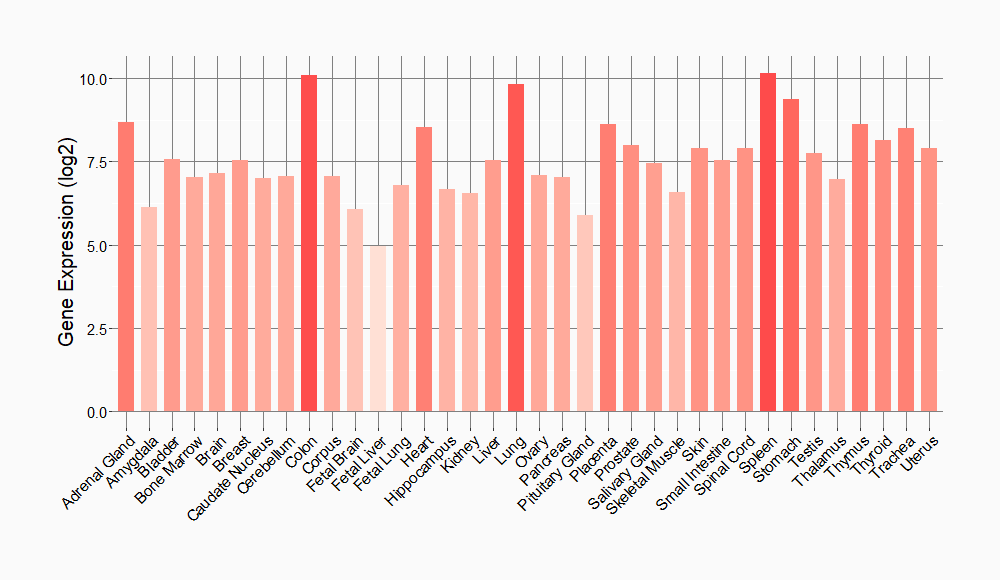
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
