Molecule Information
General Information of the Molecule (ID: Mol00085)
| Name |
High mobility group protein HMGI-C (HMGA2)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
High mobility group AT-hook protein 2; HMGIC
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
HMGA2
|
||||
| Gene ID | |||||
| Location |
chr12:65824460-65966291[+]
|
||||
| Sequence |
MSARGEGAGQPSTSAQGQPAAPAPQKRGRGRPRKQQQEPTGEPSPKRPRGRPKGSKNKSP
SKAAQKKAEATGEKRPRGRPRKWPQQVVQKKPAQEETEETSSQESAEED Click to Show/Hide
|
||||
| Function |
Functions as a transcriptional regulator. Functions in cell cycle regulation through CCNA2. Plays an important role in chromosome condensation during the meiotic G2/M transition of spermatocytes. Plays a role in postnatal myogenesis, is involved in satellite cell activation. Positively regulates IGF2 expression through PLAG1 and in a PLAG1-independent manner.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [1], [2] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-11 Fold-change: 9.89E-01 Z-score: 7.21E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | CXCR4/let-7a/HMGA2 pathway | Regulation | N.A. | |
| In Vitro Model | HPDE6-C7 cells | Pancreas | Homo sapiens (Human) | CVCL_0P38 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Transwell assay; Flow cytometric analysis | |||
| Mechanism Description | CXCR4/Let-7a axis regulates metastasis and chemoresistance of pancreatic cancer cells through targeting HMGA2. overexpression of HMGA2 restores cell proliferation, metastasis and chemosensitivity of gem inhibited by let-7a. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [3] | |||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.13E-05 Fold-change: 1.02E+00 Z-score: 4.33E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.47E-01 Fold-change: -2.87E-03 Z-score: -7.52E-02 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [7] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometric apoptosis assay | |||
| Mechanism Description | miR33b-5p sensitizes gastric cancer cells to chemotherapy drugs via inhibiting HMGA2 expression. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [8] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| HMGA2-E2F1-AKT signaling pathway | Inhibition | hsa05206 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Decreased MicroRNA-26a expression significantly decreased the expression of E2F1, diminished Akt phosphorylation, and downregulated Bcl2 expression, which causes cisplatin resistance in human non-small cell lung cancer. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [3] | |||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.13E-05 Fold-change: 1.02E+00 Z-score: 4.33E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Transwell migration assay | |||
| Mechanism Description | NEAT1 down-regulation decreased the endogenous HMGA2 expression, and HMGA2 down-regulation mimicked LncRNA NEAT1 down-regulation's effect on cell migration, invasion, EMT phenotype and chemo-sensitivity. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [6] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.56E-02 Fold-change: -2.08E-02 Z-score: -2.15E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell cycle | Activation | hsa04110 | ||
| In Vitro Model | Bel-7402/5-Fu cells | Liver | Homo sapiens (Human) | CVCL_5493 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Let-7g microRNA contributed to an increase of 5-Fu-induced cell cycle inhibit in human hepatoma cell and sensitized cells to 5-Fu, leading to increased the effectiveness of the drug in treating hepatoma cancer. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [3] | |||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.13E-05 Fold-change: 1.02E+00 Z-score: 4.33E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioma [ICD-11: 2A00.1] | [5] | |||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.12E-01 Fold-change: -6.98E-03 Z-score: -1.59E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | U87 GSCs | Brain | Homo sapiens (Human) | CVCL_0022 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-23b overexpression sensitized U87 glioma stem cells to TMZ-induced growth inhibition. And miR-23b had a synergistically suppressive effect on the expression of HMGA2 with TMZ in U87 GSCs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [7] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometric apoptosis assay | |||
| Mechanism Description | miR33b-5p sensitizes gastric cancer cells to chemotherapy drugs via inhibiting HMGA2 expression. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [4] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.12E-01; Fold-change: -2.16E-02; Z-score: -1.19E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.94E-01; Fold-change: -1.03E-02; Z-score: -1.86E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
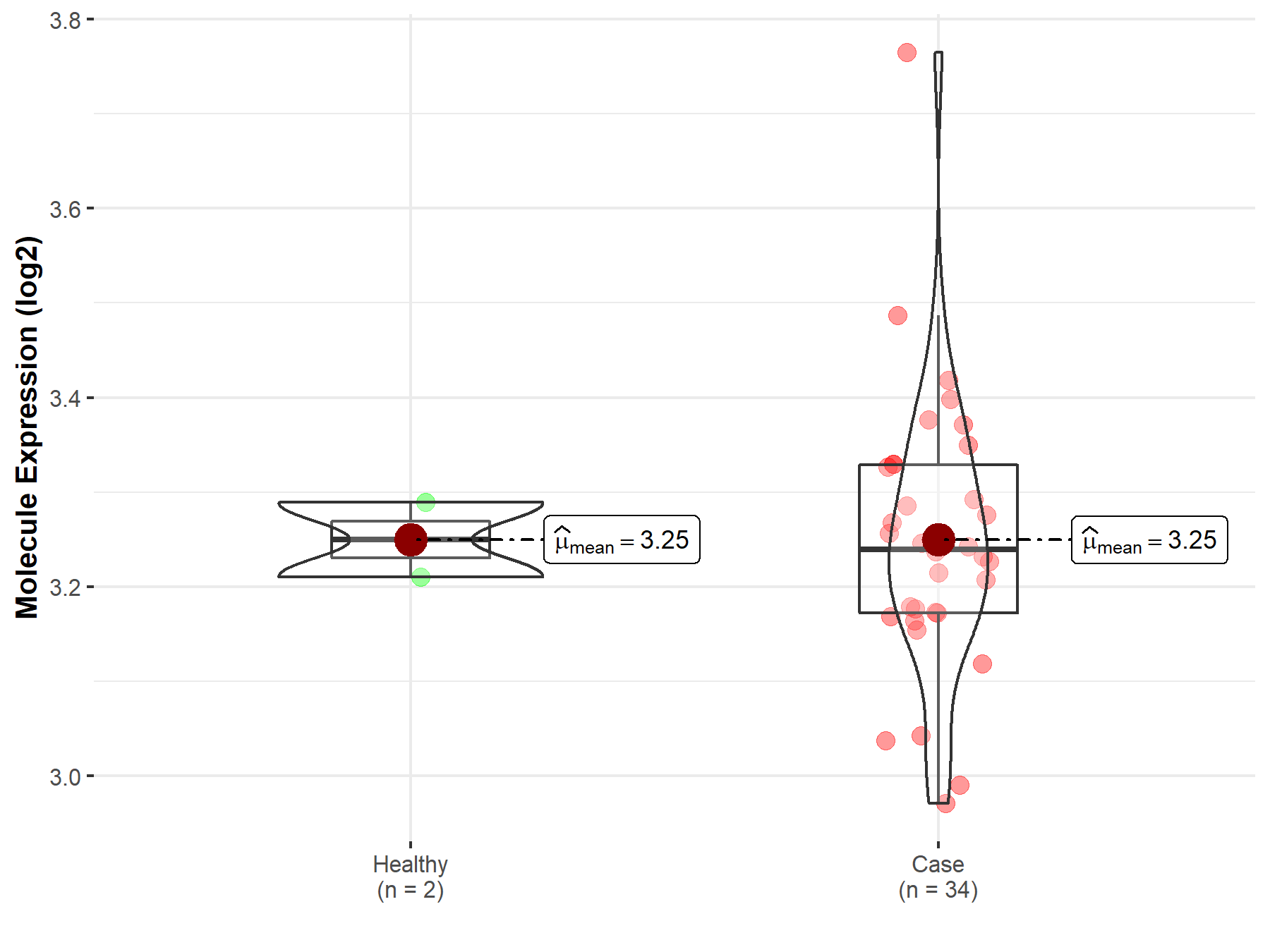
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.91E-03; Fold-change: 2.20E-01; Z-score: 9.58E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
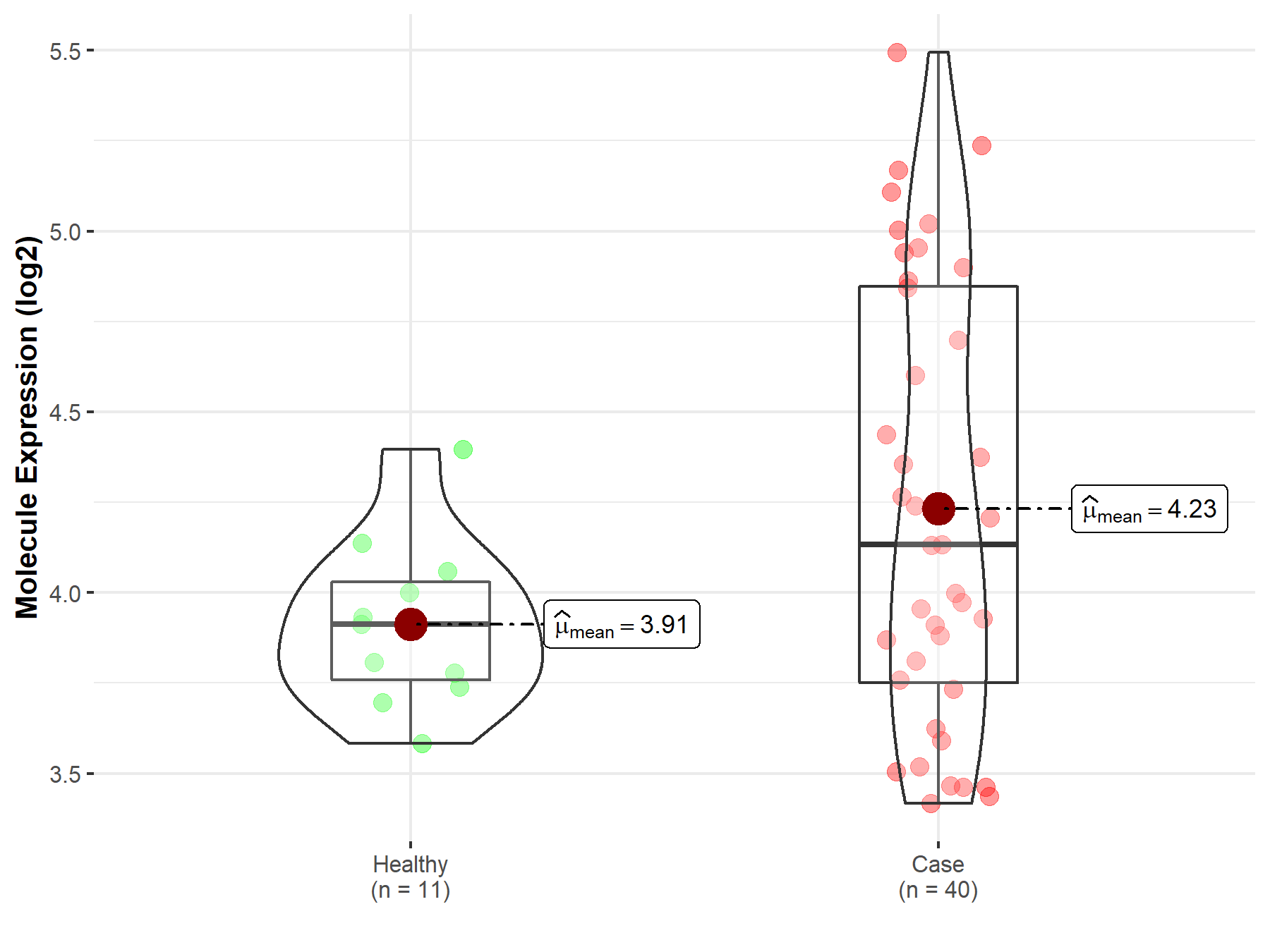
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.89E-01; Fold-change: -8.23E-02; Z-score: -4.03E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
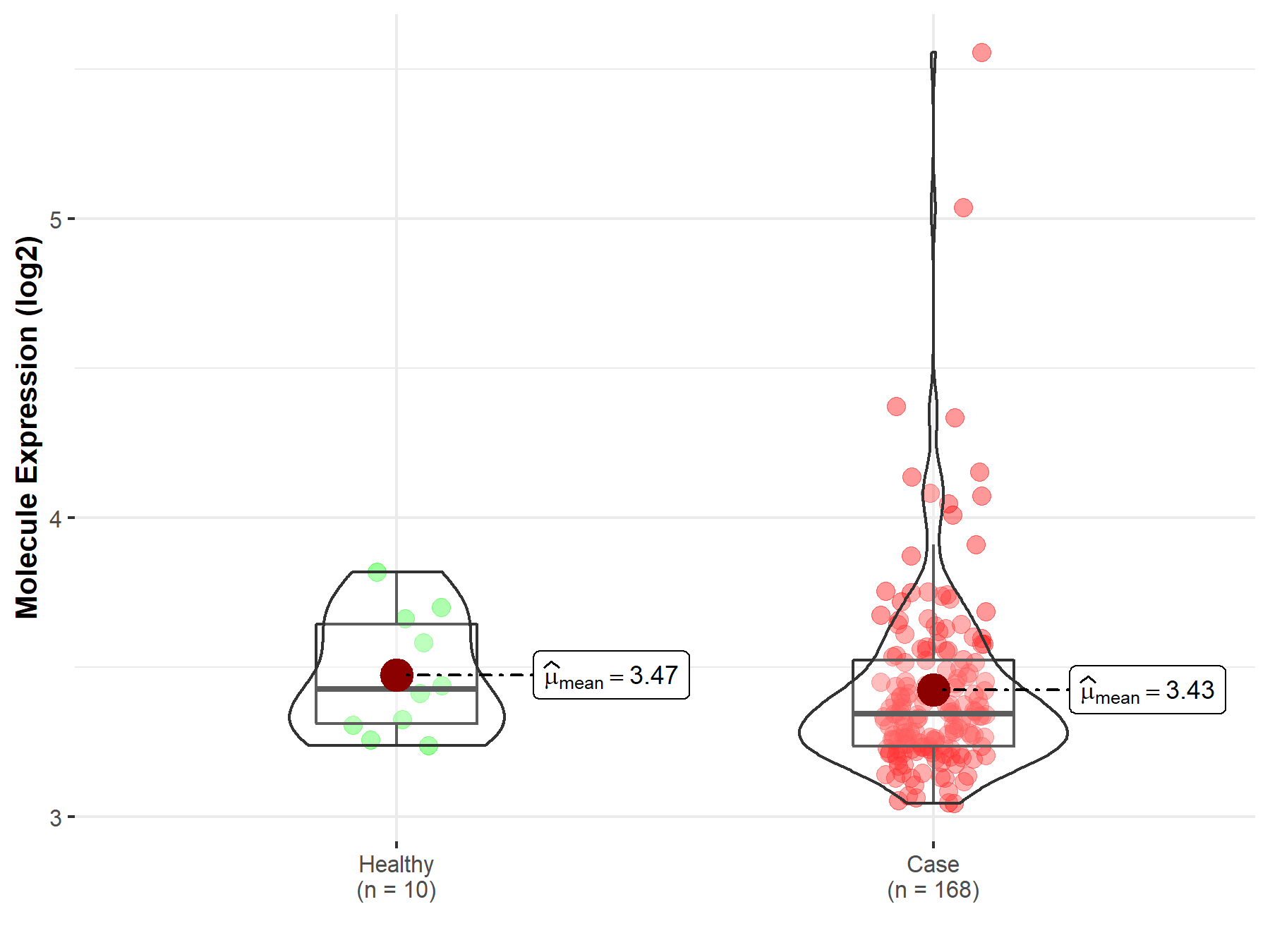
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.47E-01; Fold-change: 4.52E-02; Z-score: 3.06E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.77E-02; Fold-change: 8.29E-02; Z-score: 5.17E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
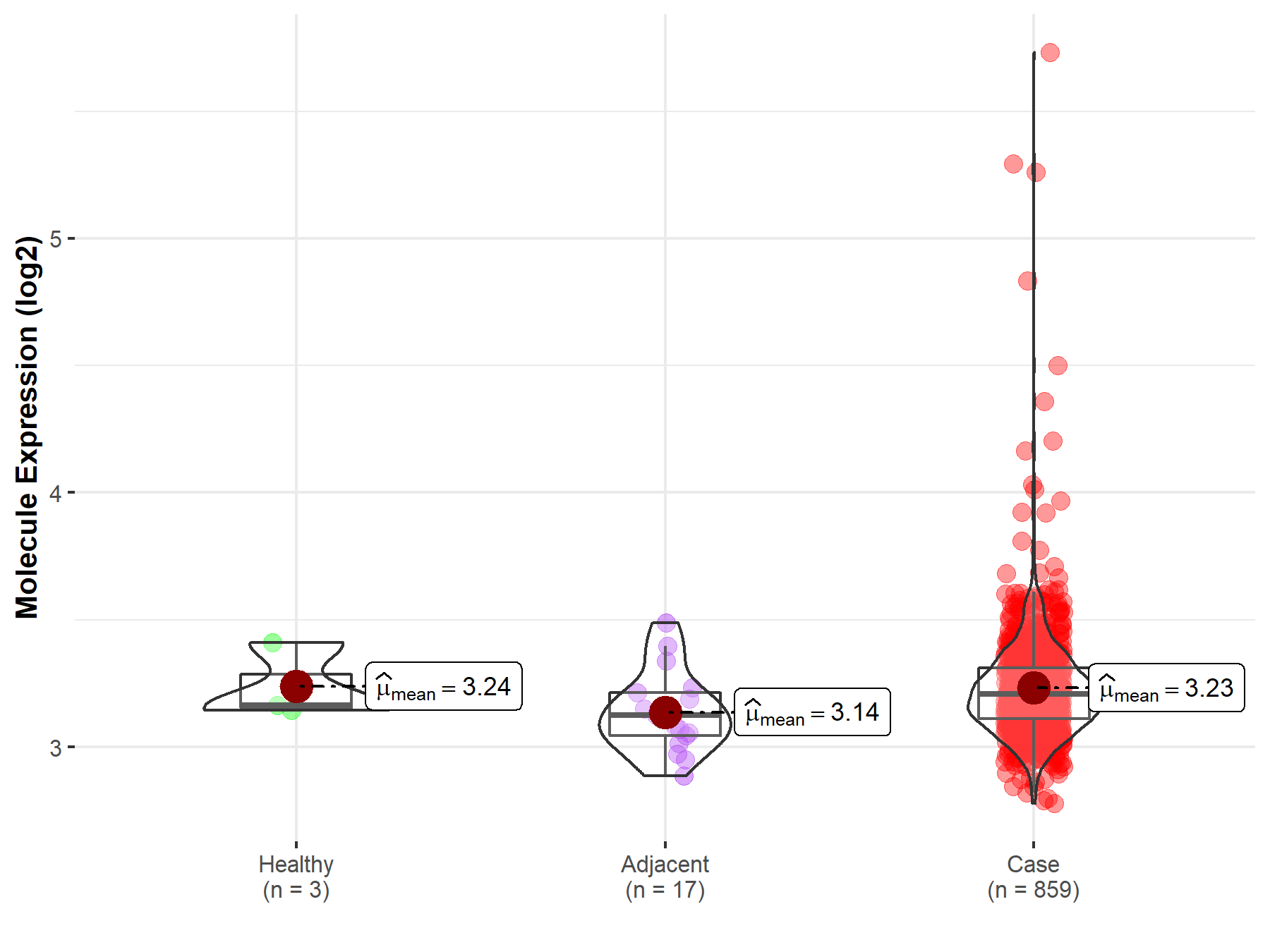
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.59E-01; Fold-change: -8.30E-02; Z-score: -4.22E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.10E-02; Fold-change: -6.78E-02; Z-score: -3.19E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
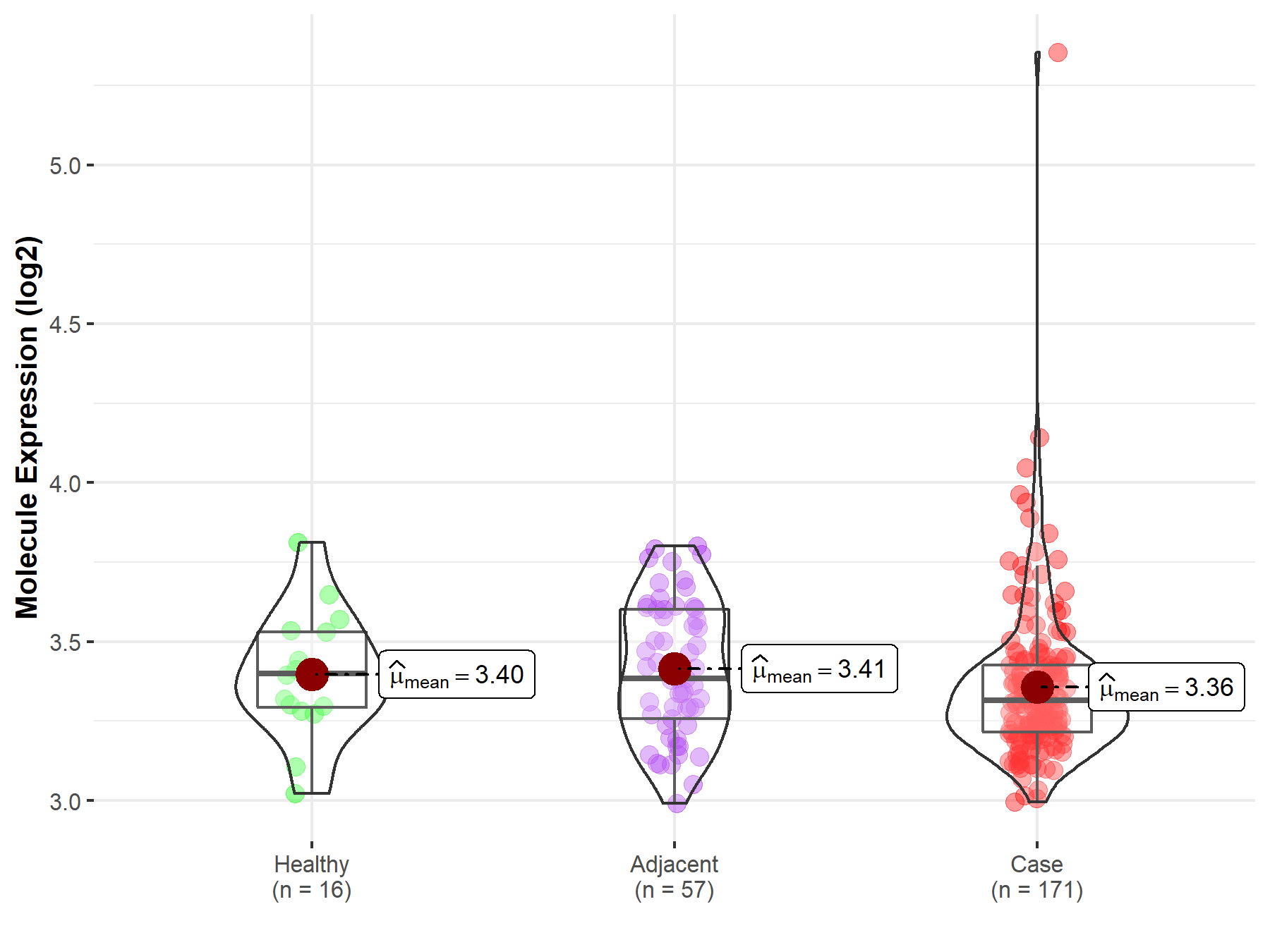
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.56E-02; Fold-change: -5.42E-02; Z-score: -4.47E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.15E-01; Fold-change: -3.04E-02; Z-score: -2.38E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 3.01E-01; Fold-change: 6.18E-03; Z-score: 6.75E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
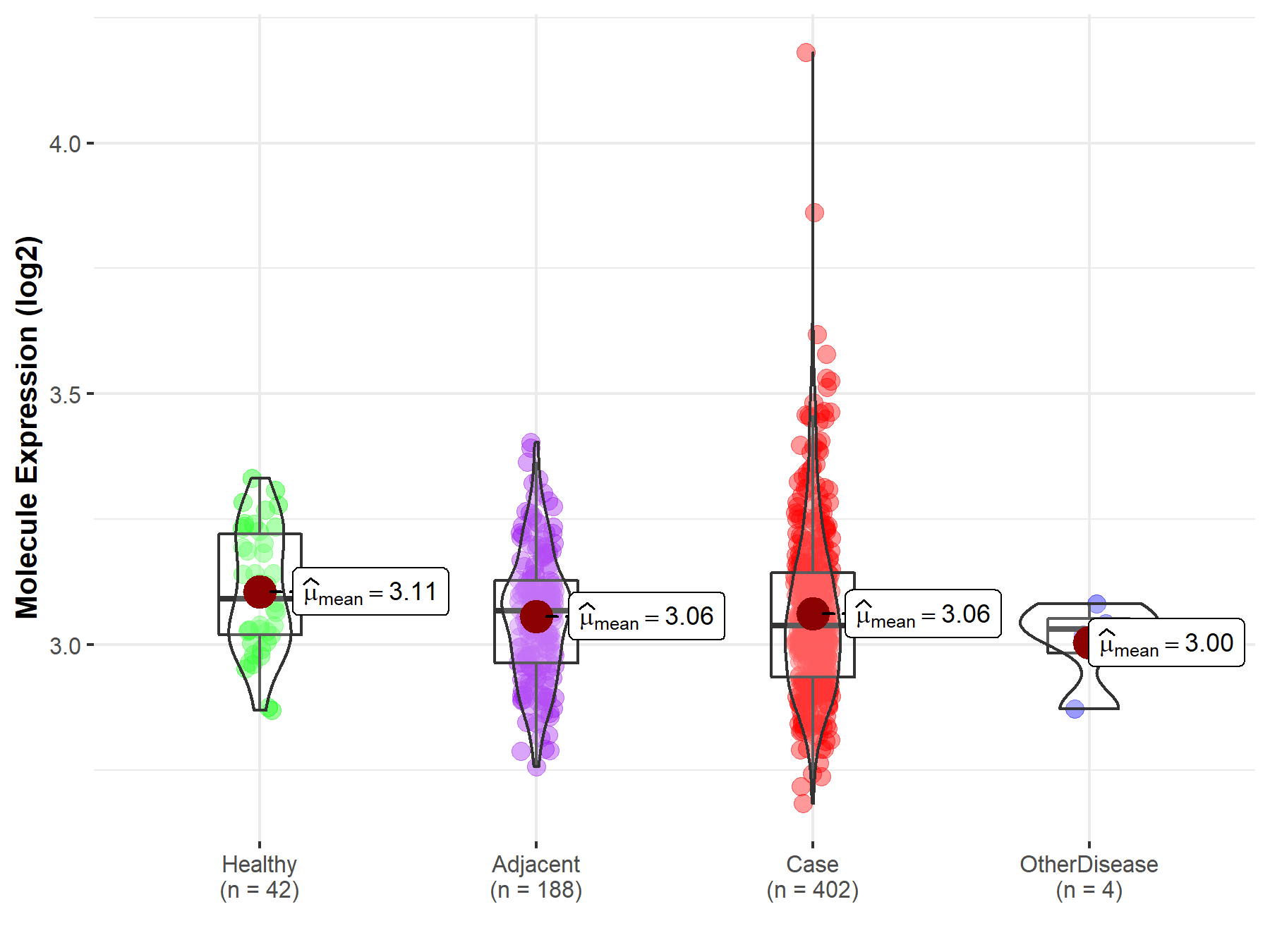
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.33E-22; Fold-change: 1.03E-01; Z-score: 8.05E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.39E-19; Fold-change: 7.90E-02; Z-score: 7.27E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
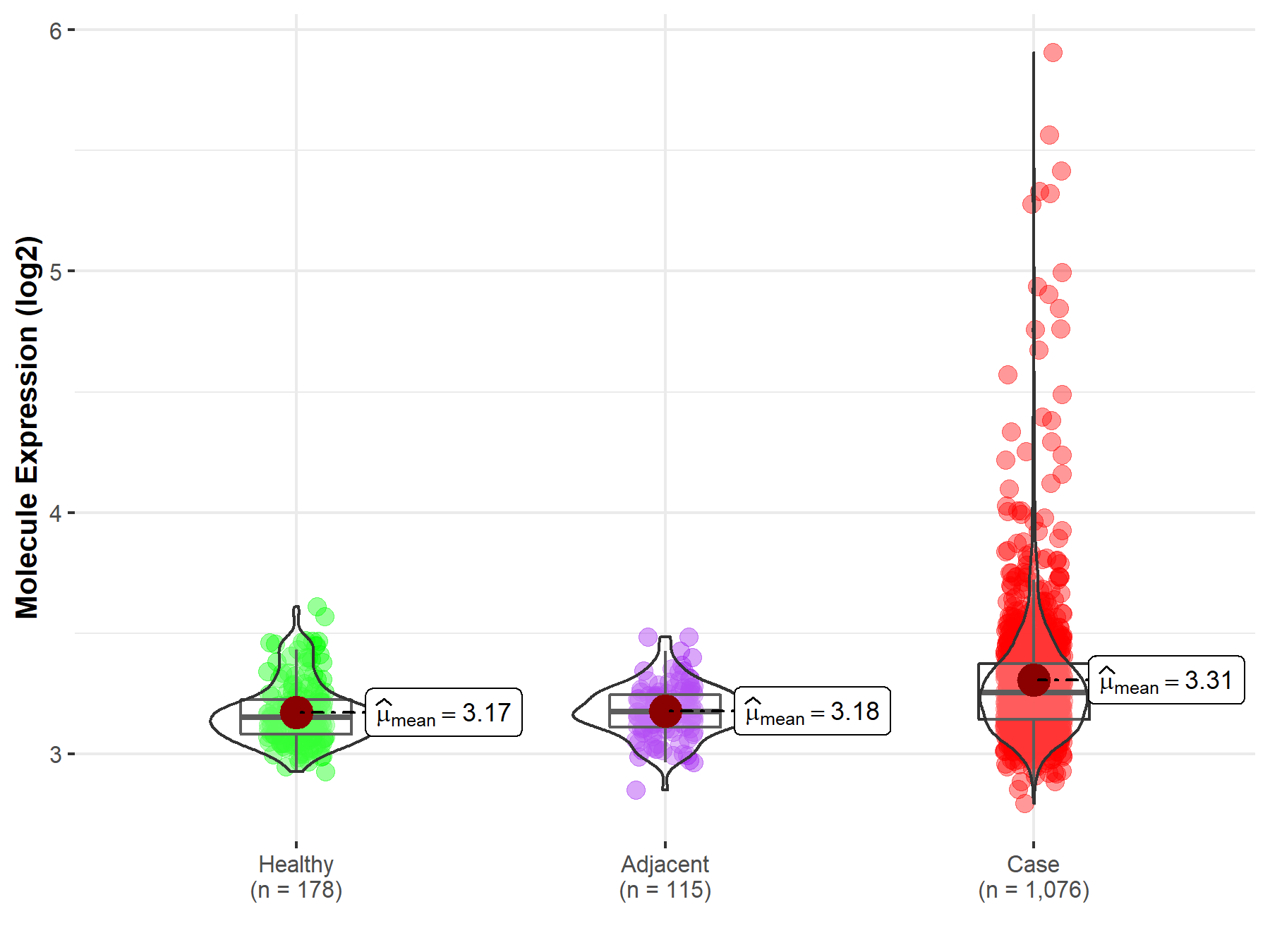
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.84E-06; Fold-change: -5.18E-02; Z-score: -2.15E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.21E-02; Fold-change: -7.54E-02; Z-score: -5.20E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
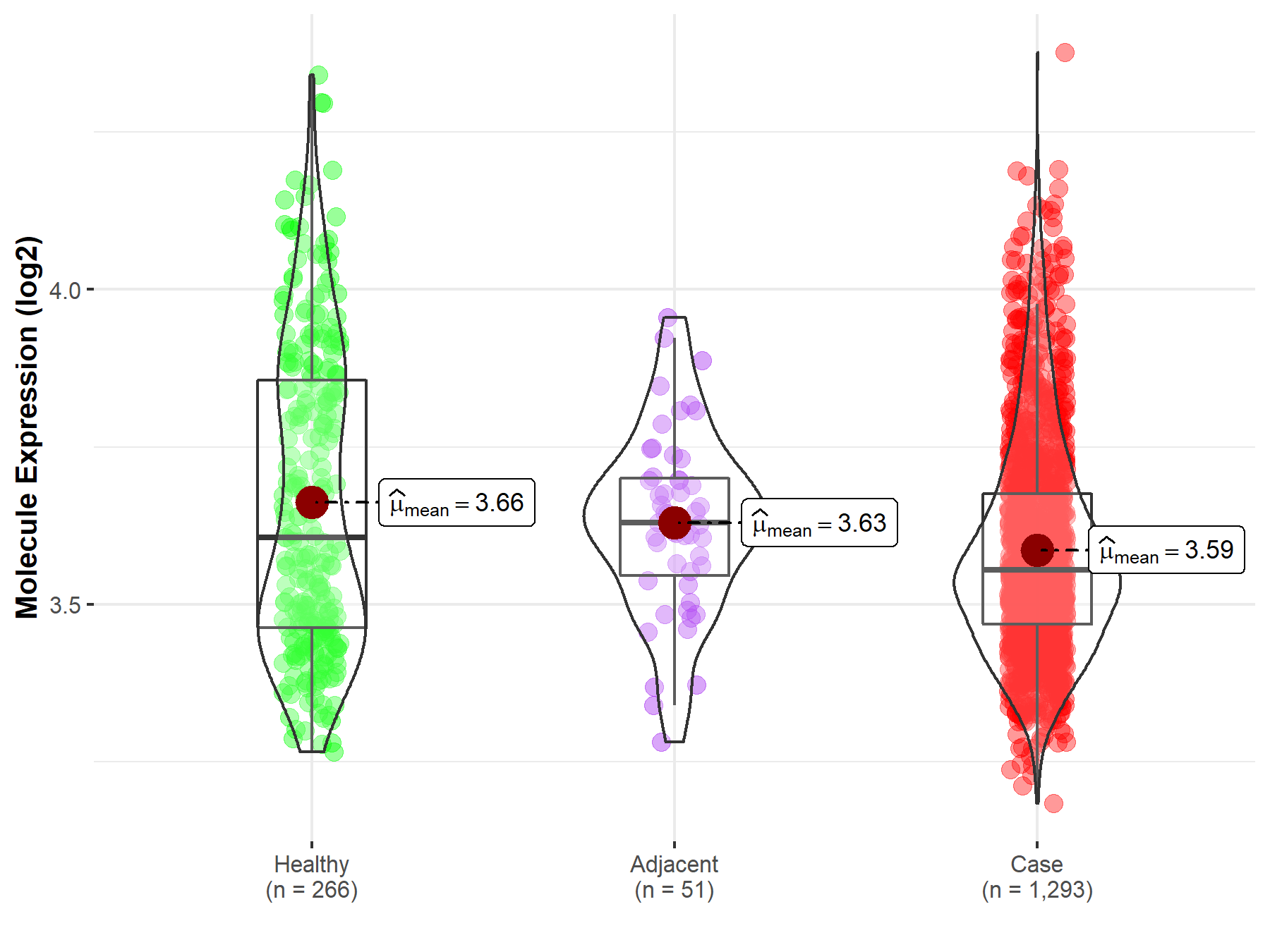
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

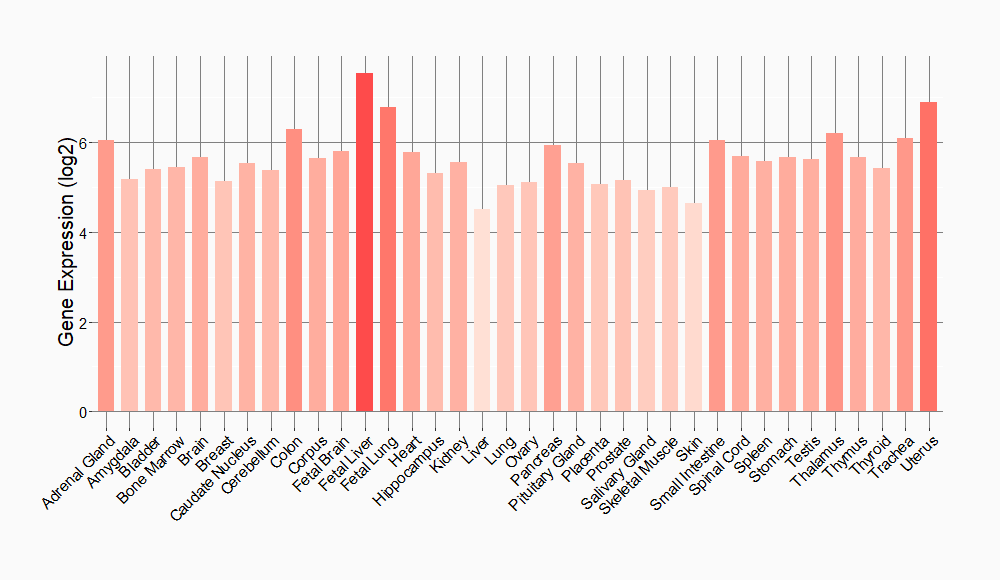
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
