Molecule Information
General Information of the Molecule (ID: Mol00035)
| Name |
Breast cancer type 1 susceptibility protein (BRCA1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
RING finger protein 53; RING-type E3 ubiquitin transferase BRCA1; RNF53
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
BRCA1
|
||||
| Gene ID | |||||
| Location |
chr17:43044295-43170245[-]
|
||||
| Sequence |
MDLSALRVEEVQNVINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQ
CPLCKNDITKRSLQESTRFSQLVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKD EVSIIQSMGYRNRAKRLLQSEPENPSLQETSLSVQLSNLGTVRTLRTKQRIQPQKTSVYI ELGSDSSEDTVNKATYCSVGDQELLQITPQGTRDEISLDSAKKAACEFSETDVTNTEHHQ PSNNDLNTTEKRAAERHPEKYQGSSVSNLHVEPCGTNTHASSLQHENSSLLLTKDRMNVE KAEFCNKSKQPGLARSQHNRWAGSKETCNDRRTPSTEKKVDLNADPLCERKEWNKQKLPC SENPRDTEDVPWITLNSSIQKVNEWFSRSDELLGSDDSHDGESESNAKVADVLDVLNEVD EYSGSSEKIDLLASDPHEALICKSERVHSKSVESNIEDKIFGKTYRKKASLPNLSHVTEN LIIGAFVTEPQIIQERPLTNKLKRKRRPTSGLHPEDFIKKADLAVQKTPEMINQGTNQTE QNGQVMNITNSGHENKTKGDSIQNEKNPNPIESLEKESAFKTKAEPISSSISNMELELNI HNSKAPKKNRLRRKSSTRHIHALELVVSRNLSPPNCTELQIDSCSSSEEIKKKKYNQMPV RHSRNLQLMEGKEPATGAKKSNKPNEQTSKRHDSDTFPELKLTNAPGSFTKCSNTSELKE FVNPSLPREEKEEKLETVKVSNNAEDPKDLMLSGERVLQTERSVESSSISLVPGTDYGTQ ESISLLEVSTLGKAKTEPNKCVSQCAAFENPKGLIHGCSKDNRNDTEGFKYPLGHEVNHS RETSIEMEESELDAQYLQNTFKVSKRQSFAPFSNPGNAEEECATFSAHSGSLKKQSPKVT FECEQKEENQGKNESNIKPVQTVNITAGFPVVGQKDKPVDNAKCSIKGGSRFCLSSQFRG NETGLITPNKHGLLQNPYRIPPLFPIKSFVKTKCKKNLLEENFEEHSMSPEREMGNENIP STVSTISRNNIRENVFKEASSSNINEVGSSTNEVGSSINEIGSSDENIQAELGRNRGPKL NAMLRLGVLQPEVYKQSLPGSNCKHPEIKKQEYEEVVQTVNTDFSPYLISDNLEQPMGSS HASQVCSETPDDLLDDGEIKEDTSFAENDIKESSAVFSKSVQKGELSRSPSPFTHTHLAQ GYRRGAKKLESSEENLSSEDEELPCFQHLLFGKVNNIPSQSTRHSTVATECLSKNTEENL LSLKNSLNDCSNQVILAKASQEHHLSEETKCSASLFSSQCSELEDLTANTNTQDPFLIGS SKQMRHQSESQGVGLSDKELVSDDEERGTGLEENNQEEQSMDSNLGEAASGCESETSVSE DCSGLSSQSDILTTQQRDTMQHNLIKLQQEMAELEAVLEQHGSQPSNSYPSIISDSSALE DLRNPEQSTSEKAVLTSQKSSEYPISQNPEGLSADKFEVSADSSTSKNKEPGVERSSPSK CPSLDDRWYMHSCSGSLQNRNYPSQEELIKVVDVEEQQLEESGPHDLTETSYLPRQDLEG TPYLESGISLFSDDPESDPSEDRAPESARVGNIPSSTSALKVPQLKVAESAQSPAAAHTT DTAGYNAMEESVSREKPELTASTERVNKRMSMVVSGLTPEEFMLVYKFARKHHITLTNLI TEETTHVVMKTDAEFVCERTLKYFLGIAGGKWVVSYFWVTQSIKERKMLNEHDFEVRGDV VNGRNHQGPKRARESQDRKIFRGLEICCYGPFTNMPTDQLEWMVQLCGASVVKELSSFTL GTGVHPIVVVQPDAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQELDTYLIPQIPH SHY Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
E3 ubiquitin-protein ligase that specifically mediates the formation of 'Lys-6'-linked polyubiquitin chains and plays a central role in DNA repair by facilitating cellular responses to DNA damage. It is unclear whether it also mediates the formation of other types of polyubiquitin chains. The BRCA1-BARD1 heterodimer coordinates a diverse range of cellular pathways such as DNA damage repair, ubiquitination and transcriptional regulation to maintain genomic stability. Regulates centrosomal microtubule nucleation. Required for appropriate cell cycle arrests after ionizing irradiation in both the S-phase and the G2 phase of the cell cycle. Required for FANCD2 targeting to sites of DNA damage. Inhibits lipid synthesis by binding to inactive phosphorylated ACACA and preventing its dephosphorylation. Contributes to homologous recombination repair (HRR) via its direct interaction with PALB2, fine-tunes recombinational repair partly through its modulatory role in the PALB2-dependent loading of BRCA2-RAD51 repair machinery at DNA breaks. Component of the BRCA1-RBBP8 complex which regulates CHEK1 activation and controls cell cycle G2/M checkpoints on DNA damage via BRCA1-mediated ubiquitination of RBBP8. Acts as a transcriptional activator.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
5 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| DNA damage repair signaling pathway | Inhibition | hsa03410 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Matrigel invasion assay | |||
| Mechanism Description | miR-638 overexpression increased sensitivity to DNA-damaging agents, ultraviolet (UV) and cisplatin. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [2] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Homologous-recombination | Regulation | N.A. | |
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| CAOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0201 | |
| C13 cells | Ovary | Homo sapiens (Human) | CVCL_0114 | |
| OV2008 cells | Ovary | Homo sapiens (Human) | CVCL_0473 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunohistochemical staining assay | |||
| Experiment for Drug Resistance |
Tumor onset measured | |||
| Mechanism Description | miR-9 bound directly to the 3'-UTR of BRCA1 and downregulated BRCA1 expression in ovarian cancer cells, miR-9 mediates the downregulation of BRCA1 and impedes DNA damage repair in ovarian cancer, improve chemotherapeutic (like cisplatin) efficacy by increasing the sensitivity of cancer cells to DNA damage. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [3] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF-7/LCC2 cells | Breast | Homo sapiens (Human) | CVCL_DP51 | |
| MECs cells | Breast | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Enforced expression of hsa-miR125a-3p in breast cancer cells potentiates docetaxel sensitivity via modulation of BRCA1 signaling. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [4] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Olaparib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| In Vivo Model | CD1 nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | miR-182-mediated down-regulation of BRCA1 impedes DNA repair, and lead to Olaparib resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [5] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Here, we describe HRD mechanisms leading to both platinum and rucaparib sensitivity (BRCA mutation, RAD51C/D alterations, and high BRCA1 promoter methylation) and summarize two important cross-resistance mechanisms: BRCA reversion mutations, and loss of BRCA1 methylation described here for the first time using archival and screening clinical specimens. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [6] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Mutations | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | UWB1.289 cells | Ovary | Homo sapiens (Human) | CVCL_B079 |
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Immunohistochemical staining assay; Multidrug resistance activity assay; Neutral comet assay | |||
| Mechanism Description | Olaparib-resistant BRCA1m OvCa cells show greater sensitivity to niraparib and rucaparib relative to other PARPis. Niraparib and rucaparib demonstrated greater cytotoxicity and reduced RF speed compared to the other three PARPis, likely due to the higher levels of SSB induction. | |||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.C61G (c.181T>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.C61G (c.181T>G) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.C64Y (c.191G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.C64Y (c.191G>A) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.M1V (c.1A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.M1V (c.1A>G) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.R71G (c.211A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.R71G (c.211A>G) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.R71K (c.212G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.R71K (c.212G>A) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.M1I (c.3G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.M1I (c.3G>T) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Nonsense | p.R1443* (c.4327C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The nonsense p.R1443* (c.4327C>T) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Nonsense | p.Q1467* (c.4399C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The nonsense p.Q1467* (c.4399C>T) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.R1495M (c.4484G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.R1495M (c.4484G>T) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.E1559K (c.4675G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.E1559K (c.4675G>A) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Rucaparib | |||
| Molecule Alteration | Missense mutation | p.D1692N (c.5074G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| Mechanism Description | The missense mutation p.D1692N (c.5074G>A) in gene BRCA1 cause the sensitivity of Rucaparib by unusual activation of pro-survival pathway | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [6] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Talazoparib | |||
| Molecule Alteration | Mutations | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | UWB1.289 cells | Ovary | Homo sapiens (Human) | CVCL_B079 |
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Immunohistochemical staining assay; Multidrug resistance activity assay; Neutral comet assay | |||
| Mechanism Description | Olaparib-resistant BRCA1m OvCa cells show greater sensitivity to niraparib and rucaparib relative to other PARPis. Niraparib and rucaparib demonstrated greater cytotoxicity and reduced RF speed compared to the other three PARPis, likely due to the higher levels of SSB induction. | |||
Clinical Trial Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [6] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Niraparib | |||
| Molecule Alteration | Mutations | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | UWB1.289 cells | Ovary | Homo sapiens (Human) | CVCL_B079 |
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Immunohistochemical staining assay; Multidrug resistance activity assay; Neutral comet assay | |||
| Mechanism Description | Olaparib-resistant BRCA1m OvCa cells show greater sensitivity to niraparib and rucaparib relative to other PARPis. Niraparib and rucaparib demonstrated greater cytotoxicity and reduced RF speed compared to the other three PARPis, likely due to the higher levels of SSB induction. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [6] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Veliparib dihydrochloride | |||
| Molecule Alteration | Mutations | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | UWB1.289 cells | Ovary | Homo sapiens (Human) | CVCL_B079 |
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Immunohistochemical staining assay; Multidrug resistance activity assay; Neutral comet assay | |||
| Mechanism Description | Olaparib-resistant BRCA1m OvCa cells show greater sensitivity to niraparib and rucaparib relative to other PARPis. Niraparib and rucaparib demonstrated greater cytotoxicity and reduced RF speed compared to the other three PARPis, likely due to the higher levels of SSB induction. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.05E-93; Fold-change: 5.27E-01; Z-score: 1.60E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.76E-19; Fold-change: 5.06E-01; Z-score: 1.86E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-05; Fold-change: 6.78E-01; Z-score: 2.89E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.38E-01; Fold-change: 3.47E-01; Z-score: 3.36E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
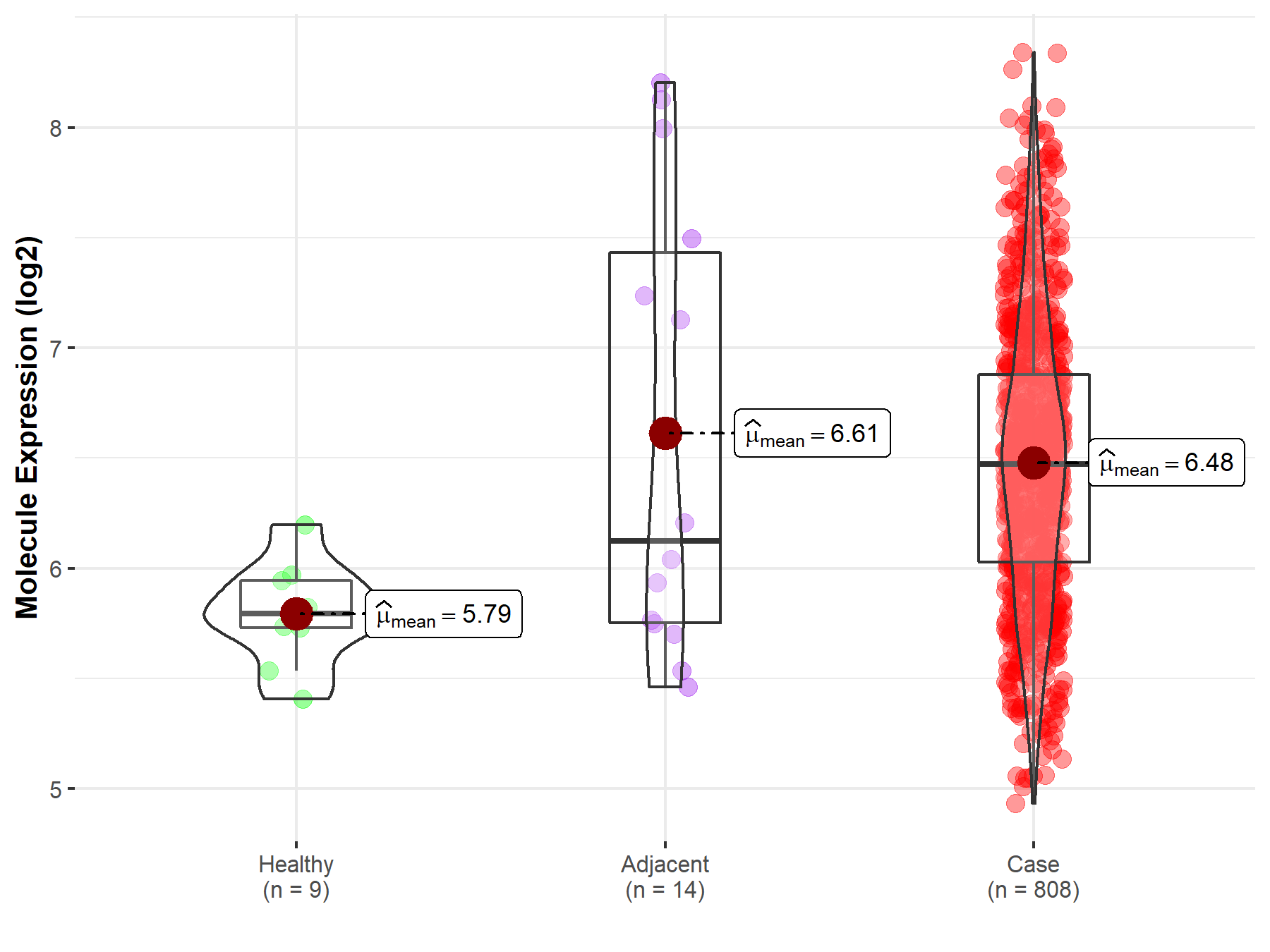
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

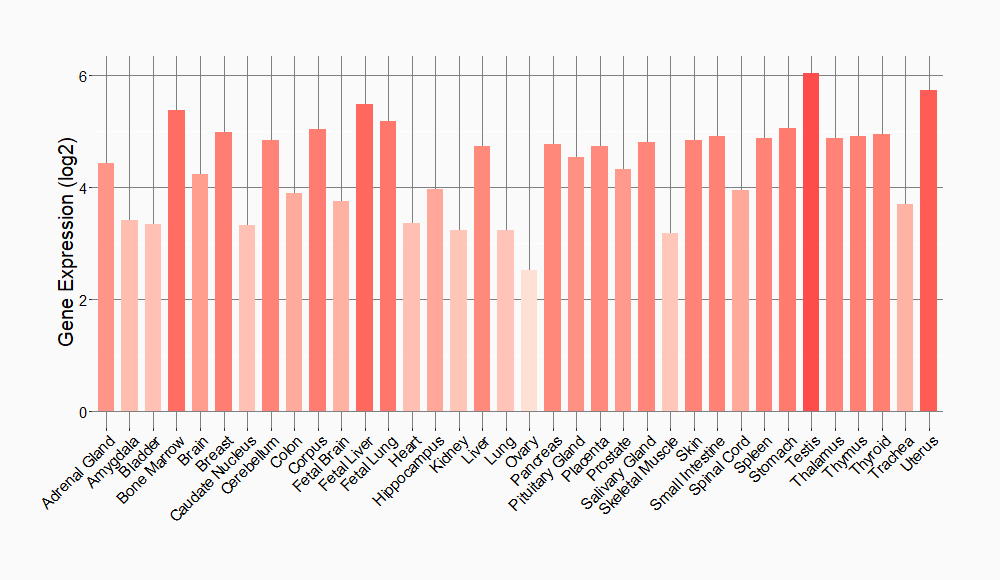
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
