Molecule Information
General Information of the Molecule (ID: Mol00017)
| Name |
Ubiquitin-like protein ATG12 (ATG12)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Autophagy-related protein 12; APG12-like; APG12; APG12L
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATG12
|
||||
| Gene ID | |||||
| Location |
chr5:115828200-115841837[-]
|
||||
| Sequence |
MAEEPQSVLQLPTSIAAGGEGLTDVSPETTTPEPPSSAAVSPGTEEPAGDTKKKIDILLK
AVGDTPIMKTKKWAVERTRTIQGLIDFIKKFLKLVASEQLFIYVNQSFAPSPDQEVGTLY ECFGSDGKLVLHYCKSQAWG Click to Show/Hide
|
||||
| Function |
Ubiquitin-like protein involved in autophagy vesicles formation. Conjugation with ATG5 through a ubiquitin-like conjugating system involving also ATG7 as an E1-like activating enzyme and ATG10 as an E2-like conjugating enzyme, is essential for its function. The ATG12-ATG5 conjugate acts as an E3-like enzyme which is required for lipidation of ATG8 family proteins and their association to the vesicle membranes.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-01 Fold-change: 9.45E-02 Z-score: 2.72E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 acts as a competing endogenous RNA for miR23b-3p and attenuates the inhibitory effect of miR23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells. MALAT1 regulates autophagy via ATG12. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/DDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR146a-5p increases chemosensitivity of NSCLC to cisplatin by targeting Atg12 to inhibit autophagy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [3] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR23b-3p/ATG12/HMGB2/autophagy regulatory loop signaling pathway | Regulation | N.A. | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | ATG12 and HMGB2 were the direct targets of miR-23b-3p. Meanwhile, ATG12 and HMGB2 were positively associated with the occurrence of autophagy. Reducing the expression of these target genes by siRNA or inhibition of autophagy both sensitized GC cells to chemotherapy. These findings suggest that a miR-23b-3p/ATG12/HMGB2/autophagy-regulatory loop has a critical role in MDR in GC. In addition, miR-23b-3p could be used as a prognostic factor for overall survival in GC. miR-23b-3p inhibited autophagy mediated by ATG12 and HMGB2 and sensitized GC cells to chemotherapy, and suggested the potential application of miR-23b-3p in drug resistance prediction and treatment. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [3] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR23b-3p/ATG12/HMGB2/autophagy regulatory loop signaling pathway | Regulation | N.A. | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | ATG12 and HMGB2 were the direct targets of miR-23b-3p. Meanwhile, ATG12 and HMGB2 were positively associated with the occurrence of autophagy. Reducing the expression of these target genes by siRNA or inhibition of autophagy both sensitized GC cells to chemotherapy. These findings suggest that a miR-23b-3p/ATG12/HMGB2/autophagy-regulatory loop has a critical role in MDR in GC. In addition, miR-23b-3p could be used as a prognostic factor for overall survival in GC. miR-23b-3p inhibited autophagy mediated by ATG12 and HMGB2 and sensitized GC cells to chemotherapy, and suggested the potential application of miR-23b-3p in drug resistance prediction and treatment. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [3] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR23b-3p/ATG12/HMGB2/autophagy regulatory loop signaling pathway | Regulation | N.A. | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | ATG12 and HMGB2 were the direct targets of miR-23b-3p. Meanwhile, ATG12 and HMGB2 were positively associated with the occurrence of autophagy. Reducing the expression of these target genes by siRNA or inhibition of autophagy both sensitized GC cells to chemotherapy. These findings suggest that a miR-23b-3p/ATG12/HMGB2/autophagy-regulatory loop has a critical role in MDR in GC. In addition, miR-23b-3p could be used as a prognostic factor for overall survival in GC. miR-23b-3p inhibited autophagy mediated by ATG12 and HMGB2 and sensitized GC cells to chemotherapy, and suggested the potential application of miR-23b-3p in drug resistance prediction and treatment. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-01; Fold-change: 3.63E-01; Z-score: 1.22E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.18E-03; Fold-change: 1.92E-01; Z-score: 5.92E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
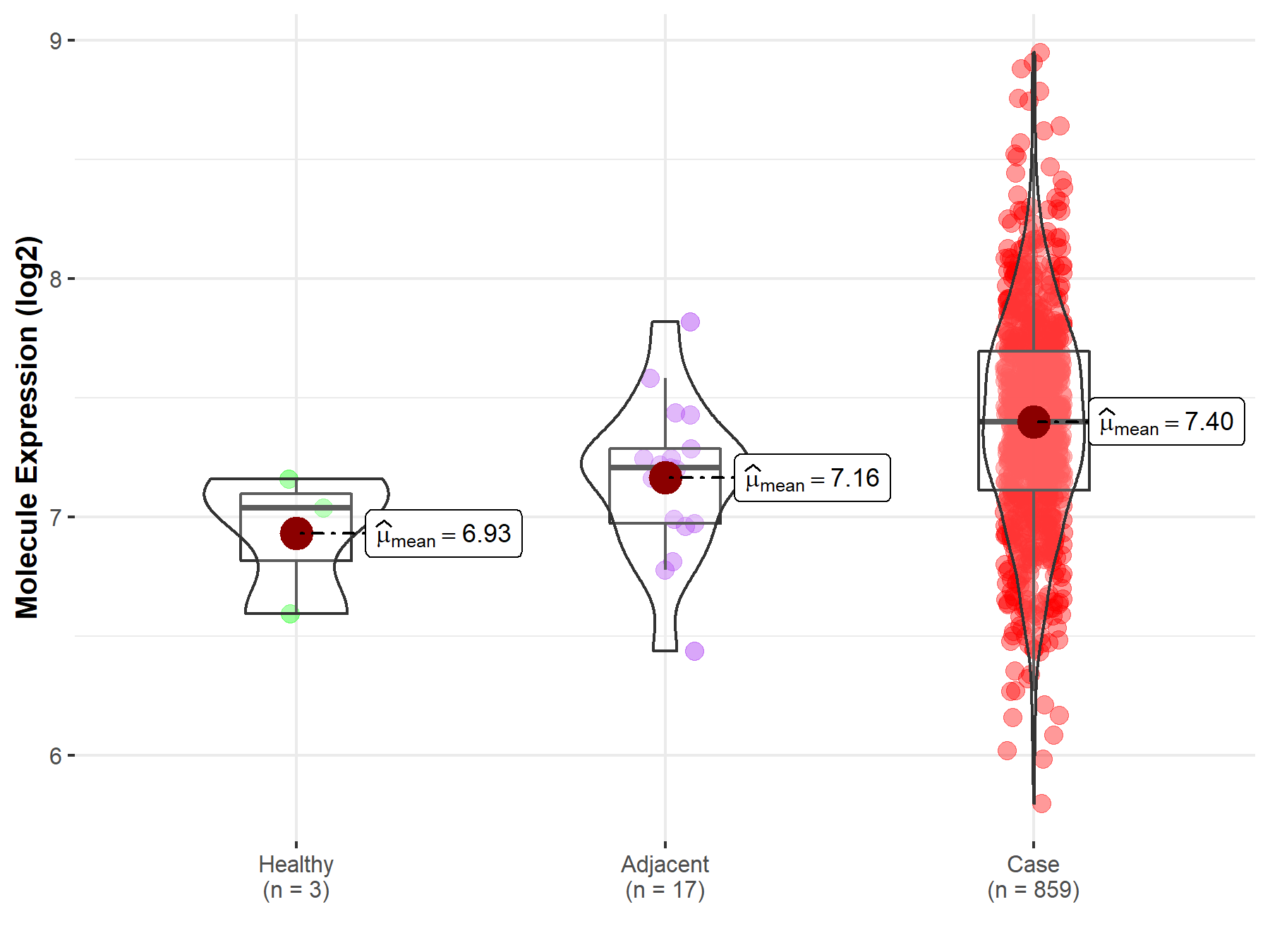
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.68E-09; Fold-change: 1.46E-01; Z-score: 4.21E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.35E-18; Fold-change: 2.93E-01; Z-score: 9.07E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

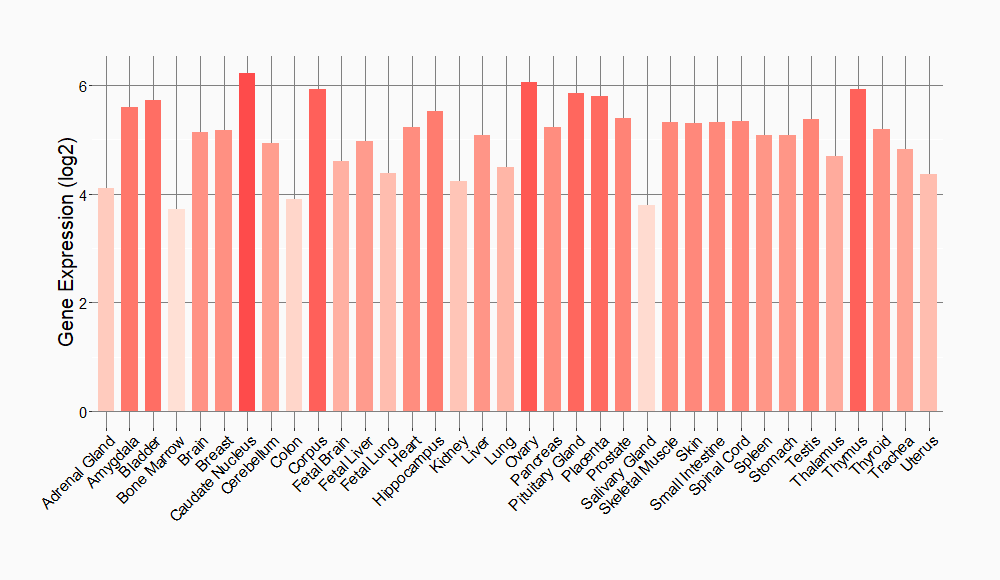
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
