Molecule Information
General Information of the Molecule (ID: Mol00013)
| Name |
Apoptotic protease-activating factor 1 (APAF1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
APAF-1; KIAA0413
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
APAF1
|
||||
| Gene ID | |||||
| Location |
chr12:98645290-98735433[+]
|
||||
| Sequence |
MDAKARNCLLQHREALEKDIKTSYIMDHMISDGFLTISEEEKVRNEPTQQQRAAMLIKMI
LKKDNDSYVSFYNALLHEGYKDLAALLHDGIPVVSSSSGKDSVSGITSYVRTVLCEGGVP QRPVVFVTRKKLVNAIQQKLSKLKGEPGWVTIHGMAGCGKSVLAAEAVRDHSLLEGCFPG GVHWVSVGKQDKSGLLMKLQNLCTRLDQDESFSQRLPLNIEEAKDRLRILMLRKHPRSLL ILDDVWDSWVLKAFDSQCQILLTTRDKSVTDSVMGPKYVVPVESSLGKEKGLEILSLFVN MKKADLPEQAHSIIKECKGSPLVVSLIGALLRDFPNRWEYYLKQLQNKQFKRIRKSSSYD YEALDEAMSISVEMLREDIKDYYTDLSILQKDVKVPTKVLCILWDMETEEVEDILQEFVN KSLLFCDRNGKSFRYYLHDLQVDFLTEKNCSQLQDLHKKIITQFQRYHQPHTLSPDQEDC MYWYNFLAYHMASAKMHKELCALMFSLDWIKAKTELVGPAHLIHEFVEYRHILDEKDCAV SENFQEFLSLNGHLLGRQPFPNIVQLGLCEPETSEVYQQAKLQAKQEVDNGMLYLEWINK KNITNLSRLVVRPHTDAVYHACFSEDGQRIASCGADKTLQVFKAETGEKLLEIKAHEDEV LCCAFSTDDRFIATCSVDKKVKIWNSMTGELVHTYDEHSEQVNCCHFTNSSHHLLLATGS SDCFLKLWDLNQKECRNTMFGHTNSVNHCRFSPDDKLLASCSADGTLKLWDATSANERKS INVKQFFLNLEDPQEDMEVIVKCCSWSADGARIMVAAKNKIFLFDIHTSGLLGEIHTGHH STIQYCDFSPQNHLAVVALSQYCVELWNTDSRSKVADCRGHLSWVHGVMFSPDGSSFLTS SDDQTIRLWETKKVCKNSAVMLKQEVDVVFQENEVMVLAVDHIRRLQLINGRTGQIDYLT EAQVSCCCLSPHLQYIAFGDENGAIEILELVNNRIFQSRFQHKKTVWHIQFTADEKTLIS SSDDAEIQVWNWQLDKCIFLRGHQETVKDFRLLKNSRLLSWSFDGTVKVWNIITGNKEKD FVCHQGTVLSCDISHDATKFSSTSADKTAKIWSFDLLLPLHELRGHNGCVRCSAFSVDST LLATGDDNGEIRIWNVSNGELLHLCAPLSEEGAATHGGWVTDLCFSPDGKMLISAGGYIK WWNVVTGESSQTFYTNGTNLKKIHVSPDFKTYVTVDNLGILYILQTLE Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Oligomeric Apaf-1 mediates the cytochrome c-dependent autocatalytic activation of pro-caspase-9 (Apaf-3), leading to the activation of caspase-3 and apoptosis. This activation requires ATP. Isoform 6 is less effective in inducing apoptosis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
5 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-05 Fold-change: -1.11E-01 Z-score: -4.51E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-221 may inhibit apoptosis by down regulating the expression of Apaf-1, so as to induce the resistance of PC-9 cells to gefitinib. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [2] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.08E-15 Fold-change: 6.49E-02 Z-score: 8.31E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Apaf-1 is a core molecule in the mitochondrial apoptotic pathway, relaying the death signal to the heptameric apoptosome complex to ignite the downstream cascade of caspases. Down-regulation of miR-155 could enhance the sensitivity of A549 cells to cisplatin treatment via restoration of the Apaf-1 pathway. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [3] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.27E-01 Fold-change: 2.19E-02 Z-score: 8.36E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Wound healing assay; Invasion assay; CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-630 inhibitor attenuated chemoresistant epithelial ovarian cancer proliferation and invasion, probably by targeting APAF-1, re-sensitizing the cells to chemotherapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [7] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||
| TGF signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| Hey A8 cells | Ovary | Homo sapiens (Human) | CVCL_8878 | |
| OVCA433 cells | Ovary | Homo sapiens (Human) | CVCL_0475 | |
| ALST cells | Ovary | Homo sapiens (Human) | CVCL_W778 | |
| HeyA8-MDR cells | Ovary | Homo sapiens (Human) | CVCL_8879 | |
| OVCA432 cells | Ovary | Homo sapiens (Human) | CVCL_3769 | |
| OVCAR5 cells | Ovary | Homo sapiens (Human) | CVCL_1628 | |
| SkOV3-TR cells | Ovary | Homo sapiens (Human) | CVCL_HF69 | |
| SkOV3ip cells | Ovary | Homo sapiens (Human) | CVCL_0C84 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Flow cytometric assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The identification of APAF1 as a direct target of miR21 and APAF1 as a mediator of miR21 for conferring chemoresistance in ovarian cancer suggests that strategies based on the upregulation of APAF1 in ovarian cancer cells can be used to sensitize ovarian cancer cells to paclitaxel treatment.ovarian cancer suggests that strategies based on the upregulation of APAF1 in ovarian cancer cells can be used to sensitize ovarian cancer cells to paclitaxel treatment. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute promyelocytic leukemia [ICD-11: 2A60.2] | [5] | |||
| Resistant Disease | Acute promyelocytic leukemia [ICD-11: 2A60.2] | |||
| Resistant Drug | Arsenic trioxide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NB4 cells | Bone marrow | Homo sapiens (Human) | CVCL_0005 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | ATO-resistant APL cells showed upregulation of?APAF1,?BCL2,?BIRC3, and?NOL3?genes, while?CD70?and?IL10?genes were downregulated, compared to ATO-sensitive cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [6] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | APAF-1/caspase-9 apoptotic signaling pathway | Activation | hsa04210 | |
| Cell apoptosis | Inhibition | hsa04210 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-23a may inhibit 5-FU-induced apoptosis through the APAF-1/caspase-9 pathway and provide new insight into CRC treatment. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [8] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | TRAIL | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| FHC cells | Colon | Homo sapiens (Human) | CVCL_3688 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | Knockdown of miR27a sensitizes colorectal cancer stem cells to TRAIL by promoting the formation of Apaf-1-caspase-9 complex. miR27a antioligonucleotides enhanced the anti-tumor effect of TRAIL on colorectal cancer stem cells via increasing the expression of Apaf-1. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.08E-15; Fold-change: 1.71E-01; Z-score: 4.75E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.36E-01; Fold-change: 6.53E-02; Z-score: 2.03E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
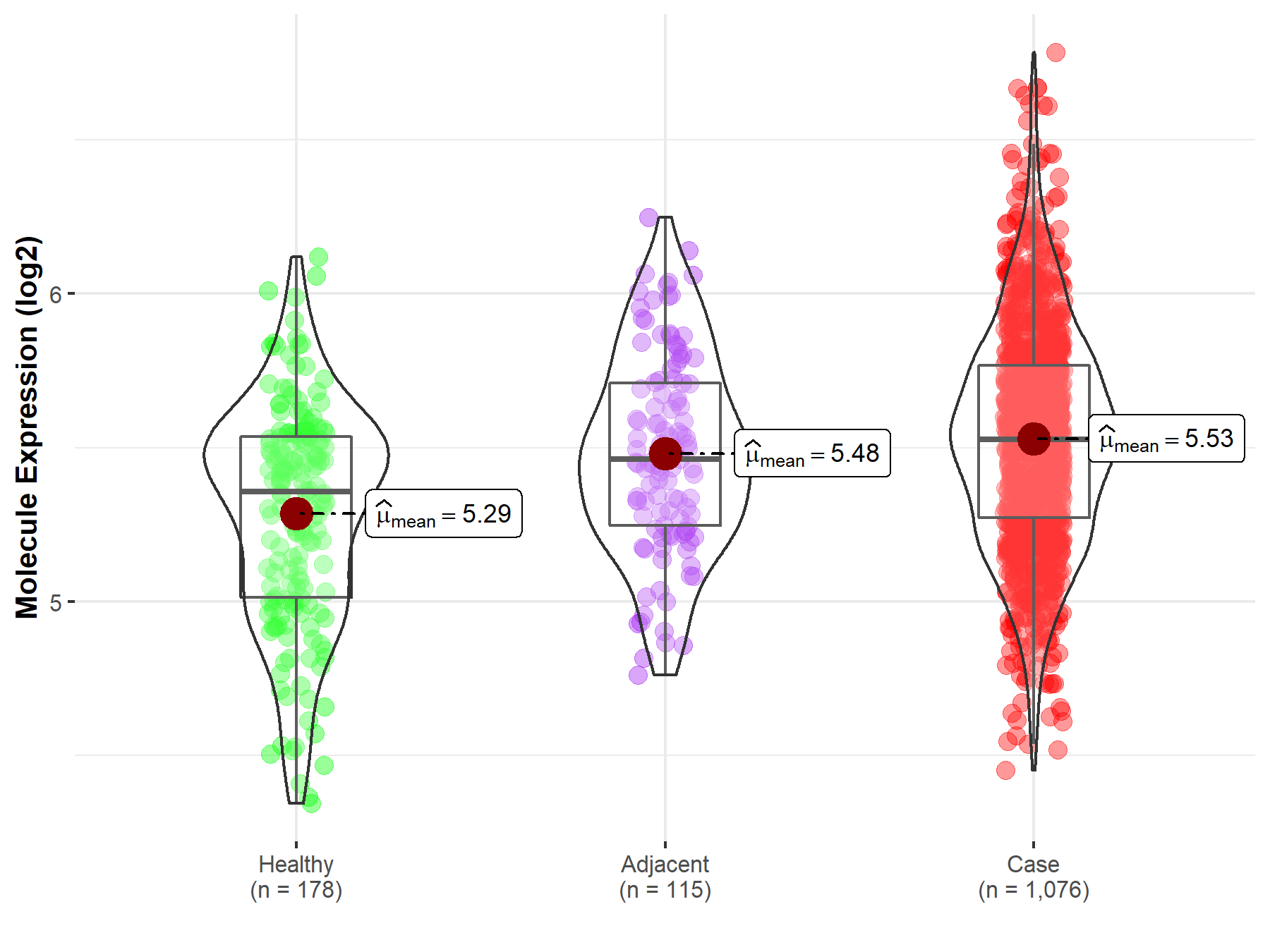
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.27E-01; Fold-change: 4.51E-02; Z-score: 1.51E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.04E-01; Fold-change: -1.63E-01; Z-score: -3.28E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
