Drug Information
Drug (ID: DG00232) and It's Reported Resistant Information
| Name |
Piperaquine
|
||||
|---|---|---|---|---|---|
| Synonyms |
Piperaquinoline; Quinoline, 4,4'-(1,3-propanediyldi-4,1-piperazinediyl)bis(7-chloro-); Quinoline, 4,4'-(1,3-propanediyldi-4,1-piperazinediyl)bis(7-chloro-(9CI); 1,3-bis(1-(7-chloro-4'-quinolyl)-4'-piperazinyl)propane; 1,3-bis(4-(7'-chloro-4'-quinoline)-1-piperazine); 4,4'-(propane-1,3-diyldipiperazine-4,1-diyl)bis(7-chloroquinoline); 7-chloro-4-[4-[3-[4-(7-chloroquinolin-4-yl)piperazin-1-yl]propyl]piperazin-1-yl]quinoline
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
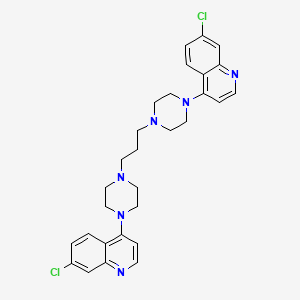
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[5]
|
||||
| Target | Sarcoplasmic/endoplasmic reticulum calcium ATPase (ATP2A) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C29H32Cl2N6
|
||||
| IsoSMILES |
C1CN(CCN1CCCN2CCN(CC2)C3=C4C=CC(=CC4=NC=C3)Cl)C5=C6C=CC(=CC6=NC=C5)Cl
|
||||
| InChI |
1S/C29H32Cl2N6/c30-22-2-4-24-26(20-22)32-8-6-28(24)36-16-12-34(13-17-36)10-1-11-35-14-18-37(19-15-35)29-7-9-33-27-21-23(31)3-5-25(27)29/h2-9,20-21H,1,10-19H2
|
||||
| InChIKey |
UCRHFBCYFMIWHC-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloroquine resistance transporter (CRT) | [1], [2], [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.H97Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | In contrast, gene-edited parasites with PfCRT H97Y, F145I, M343L, or G353V mutations are resistant to piperaquine in vitro. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [1], [4] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.F145I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | In contrast, gene-edited parasites with PfCRT H97Y, F145I, M343L, or G353V mutations are resistant to piperaquine in vitro. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [1], [4] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.G353V |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | In contrast, gene-edited parasites with PfCRT H97Y, F145I, M343L, or G353V mutations are resistant to piperaquine in vitro. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [1], [4] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.M343L |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | In contrast, gene-edited parasites with PfCRT H97Y, F145I, M343L, or G353V mutations are resistant to piperaquine in vitro. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.F145I + p.G353V+ p.I218F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
PacBio amplicon sequencing assay; Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Piperaquine susceptibility testing assay | |||
| Mechanism Description | In parasites with single-copy pfpm2, those with the PfCRT F145I, G353V, or I218F mutations had a significantly greater log10-transformed piperaquine IC90 compared to Dd2 (linear regression; P <.0001, P =.0022, and P =.019, respectively), while other mutations did not show a significant difference in piperaquine IC90 compared to Dd2 (perhaps owing to smaller sample. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation + Chromosome variation | PfCRT p.F145I+p.G353V+p.I218F + Haplotype |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
PacBio amplicon sequencing assay; Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Piperaquine susceptibility testing assay | |||
| Mechanism Description | Parasites with the Dd2 haplotype and pfpm2 amplification had significantly greater mean log10-transformed piperaquine IC90 compared to Dd2 parasites without pfpm2 amplification (t test, P =.0079). In parasites with newly emerged PfCRT mutations, mean log10-transformed piperaquine IC90 was not significantly different between parasites with or without pfpm2 amplification. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [6] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.T93S+p.H97Y+p.F145I+p.I218F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Piperaquine survival assay | |||
| Mechanism Description | The characterization of culture-adapted isolates revealed that the presence of novel pfcrt mutations (T93S, H97Y, F145I, and I218F) with E415G-Exo mutation can confer PPQ-resistance, in the absence of pfpm2 amplification. | |||
| Key Molecule: Putative chloroquine resistance transporter (PVCRT) | [7] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C350R |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Yeast strain CH1305 | 4932 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | At 300 uM PPQ, C350R/7G8 PfCRT shows a 3.9-fold increased rate of PPQ transport relative to that of 7G8 PfCRT, and F145I/Dd2 PfCRT shows a 2.7-fold increased rate of PPQ transport relative to that of Dd2. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [5] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum asexual blood-stage parasites | 5833 | ||
| Experiment for Molecule Alteration |
DNA clones asssay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Addition of the C101F mutation to the chloroquine (CQ) resistance-conferring PfCRT Dd2 isoform common to Asia can confer PPQ resistance to cultured parasites. Resistance was demonstrated as significantly higher PPQ concentrations causing 90% inhibition of parasite growth (IC90) or 50% parasite killing. | |||
|
|
||||
| Key Molecule: Plasmepsin II (PMII) | [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation + Chromosome variation | PfCRT p.F145I+p.G353V+p.I218F + pfpm2 Amplification |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
PacBio amplicon sequencing assay; Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Piperaquine susceptibility testing assay | |||
| Mechanism Description | Parasites with the Dd2 haplotype and pfpm2 amplification had significantly greater mean log10-transformed piperaquine IC90 compared to Dd2 parasites without pfpm2 amplification (t test, P =.0079). In parasites with newly emerged PfCRT mutations, mean log10-transformed piperaquine IC90 was not significantly different between parasites with or without pfpm2 amplification. | |||
| Key Molecule: Plasmepsin II (PMII) | [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Chromosome variation | pfpm2 Amplification+Haplotype |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
PacBio amplicon sequencing assay; Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Piperaquine susceptibility testing assay | |||
| Mechanism Description | Parasites with the Dd2 haplotype and pfpm2 amplification had significantly greater mean log10-transformed piperaquine IC90 compared to Dd2 parasites without pfpm2 amplification (t test, P?=?.0079). | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
