Drug Information
Drug (ID: DG00077) and It's Reported Resistant Information
| Name |
Lumefantrine
|
||||
|---|---|---|---|---|---|
| Synonyms |
Benflumetol; Coartem; Dl-Benflumelol; Lumefantrine (INN); Lumefantrine [INN:BAN]; (+-)-2,7-Dichloro-9-((Z)-p-chlorobenzylidene)-alpha((dibutylamino)methyl)fluorene-4-methanol; (+-)-2,7-Dichloro-9-((Z)-p-chlorobenzylidene)-alpha-((dibutylamino)methyl)fluorene-4-methanol; 2-(dibutylamino)-1-[(9Z)-2,7-dichloro-9-(4-chlorobenzylidene)-9H-fluoren-4-yl]ethanol; 2-(dibutylamino)-1-[(9Z)-2,7-dichloro-9-[(4-chlorophenyl)methylidene]fluoren-4-yl]ethanol; 2-Dibutylamino-1-[2,7-dichloro-9-(4-chloro-benzylidene)-9H-fluoren-4-yl]-ethanol; 2-Dibutylamino-1-{2,7-dichloro-9-[1-(4-chloro-phenyl)-meth-(Z)-ylidene]-9H-fluoren-4-yl}-ethanol
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
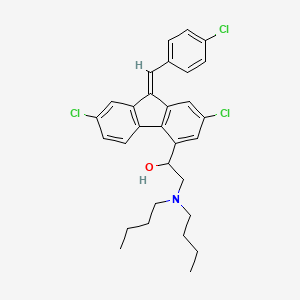
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[2]
|
||||
| Target | Sodium pump subunit alpha-1 (ATP1A1) | AT1A1_HUMAN | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C30H32Cl3NO
|
||||
| IsoSMILES |
CCCCN(CCCC)CC(C1=CC(=CC\\2=C1C3=C(/C2=C/C4=CC=C(C=C4)Cl)C=C(C=C3)Cl)Cl)O
|
||||
| InChI |
1S/C30H32Cl3NO/c1-3-5-13-34(14-6-4-2)19-29(35)28-18-23(33)17-27-25(15-20-7-9-21(31)10-8-20)26-16-22(32)11-12-24(26)30(27)28/h7-12,15-18,29,35H,3-6,13-14,19H2,1-2H3/b25-15-
|
||||
| InChIKey |
DYLGFOYVTXJFJP-MYYYXRDXSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloroquine resistance transporter (CRT) | [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N326S+p.I356T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Quantitative trait loci (QTL) assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Comparisons of the MEF and HLF responses showed that the Cambodian 803 line, as for LUM, was less susceptible than Ghanaian GB4 to these drugs: the geometric mean EC50s of 803 relative to GB4 were 2.9-fold greater with MEF and 4.6-fold greater with HLF, whereas these were 2.0-fold greater with CQ and 1.7-fold reduced w. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Drug Resistance |
HRP-2 ELISA assay | |||
| Mechanism Description | Isolates with multiple pfmdr1 copies had significantly higher IC50s against OZ78, OZ277, MQ, and LUM. In contrast, no significant differences in IC50s between isolates with single and multiple pfmdr1 copy numbers were observed for the other test compounds. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [1] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N86Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Despite the availability of few mutant parasites for comparison, the PfMDR1 Asn86Tyr substitution appeared to be associated with increased susceptibility to lumefantrine and mefloquine, as seen prev. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [3] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N86+p.Y184 |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum isolates | 5833 | ||
| Experiment for Molecule Alteration |
Quantitative trait loci (QTL) assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | The geometric mean LUM EC50 of 803 was 5.8-fold greater than GB4 (3.21 nM, 95% Confidence Interval 2.80-3.66 nM vs. 0.55 nM, 95% CI 0.46-0.67 nM, respectively). The Cambodian 803 line, as for LUM, was less susceptible than Ghanaian GB4 to these drugs: the geometric mean EC50s of 803 relative to GB4 were 2.9-fold greater with MEF and 4.6-fold greater with HLF, whereas these were 2.0-fold greater with CQ and 1.7-fold reduced with DHA. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [4] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K76T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Nested PCR | |||
| Mechanism Description | Both in vitro and molecular surveillance studies have associated CQ resistance mainly with the pfcrt 76T allele, but also with pfmdr1 86Y and 184F alleles. Pfcrt 76T and pfmdr1 86Y mutant alleles have also been reported to decrease P. falciparum susceptibility to amodiaquine but increase parasite sensitivity to dihydroartemisinin, lumefantrine and mefl. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [4] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N86Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Nested PCR | |||
| Mechanism Description | Both in vitro and molecular surveillance studies have associated CQ resistance mainly with the pfcrt 76T allele, but also with pfmdr1 86Y and 184F alleles. Pfcrt 76T and pfmdr1 86Y mutant alleles have also been reported to decrease P. falciparum susceptibility to amodiaquine but increase parasite sensitivity to dihydroartemisinin, lumefantrine and mefl. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
